|
|
|
|
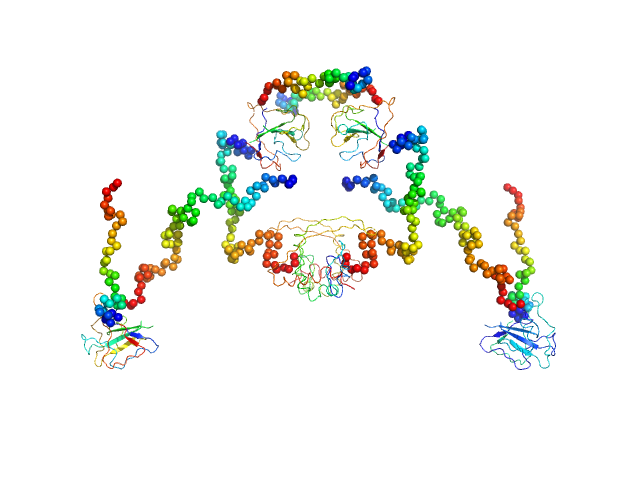
|
| Sample: |
Nucleoprotein tetramer, 183 kDa Middle East respiratory … protein
5-(2-methoxyethoxy)-1H-indole tetramer, 1 kDa
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2019 Nov 26
|
Targeting the N-Terminus Domain of the Coronavirus Nucleocapsid Protein Induces Abnormal Oligomerization via Allosteric Modulation
Frontiers in Molecular Biosciences 9 (2022)
Hsu J, Chen J, Lin S, Hong J, Chen Y, Jeng U, Luo S, Hou M
|
| RgGuinier |
6.0 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
500 |
nm3 |
|
|
|
|
|
|
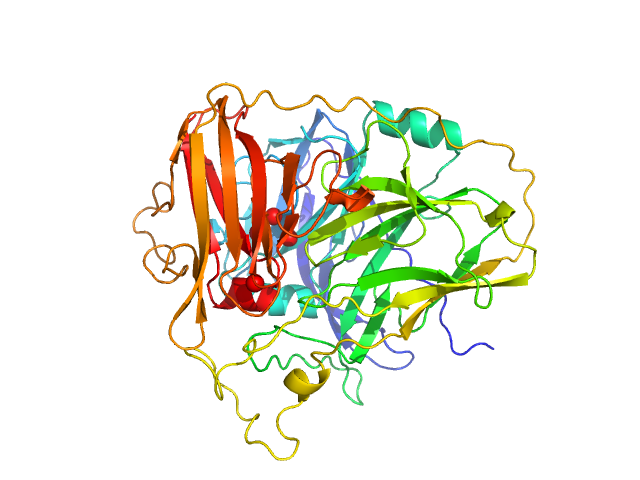
|
| Sample: |
McoA evolved variant 2F4 (Periplasmic cell division protein (SufI)) monomer, 55 kDa Aquifex aeolicus VF5 protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 25
|
Distal Mutations Shape Substrate-Binding Sites during Evolution of a Metallo-Oxidase into a Laccase
ACS Catalysis :5022-5035 (2022)
Brissos V, Borges P, Núñez-Franco R, Lucas M, Frazão C, Monza E, Masgrau L, Cordeiro T, Martins L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aquifex aeolicus McoA metaloxidase ∆328-352 evolved variant (2F4∆328-352) monomer, 53 kDa Aquifex aeolicus protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 18
|
Distal Mutations Shape Substrate-Binding Sites during Evolution of a Metallo-Oxidase into a Laccase
ACS Catalysis :5022-5035 (2022)
Brissos V, Borges P, Núñez-Franco R, Lucas M, Frazão C, Monza E, Masgrau L, Cordeiro T, Martins L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
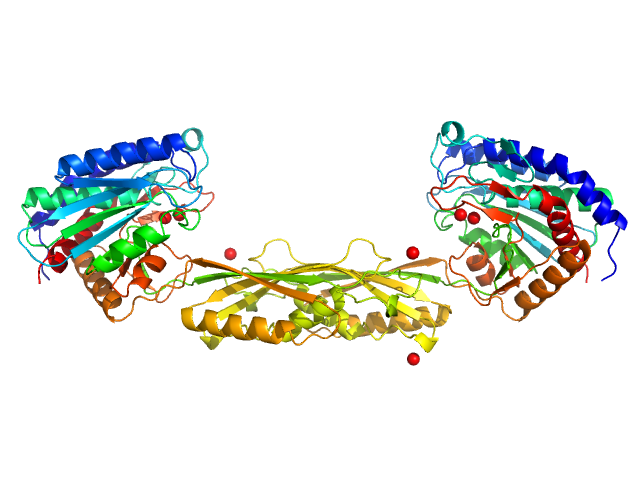
|
| Sample: |
Carboxypeptidase G2 (circular permutant CP-N89) K177A dimer, 85 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jun 6
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
|
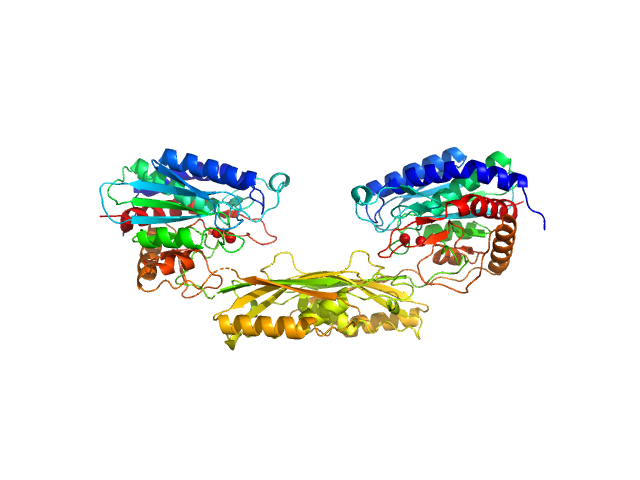
| Sample: |
Carboxypeptidase G2 (circular permutant CP-N89) K177A dimer, 85 kDa Pseudomonas sp. (strain … protein
Methotrexate dimer, 1 kDa
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jun 6
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|
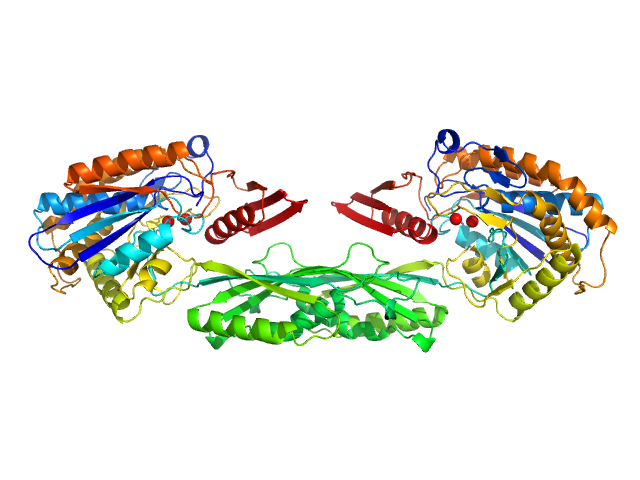
|
| Sample: |
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
|
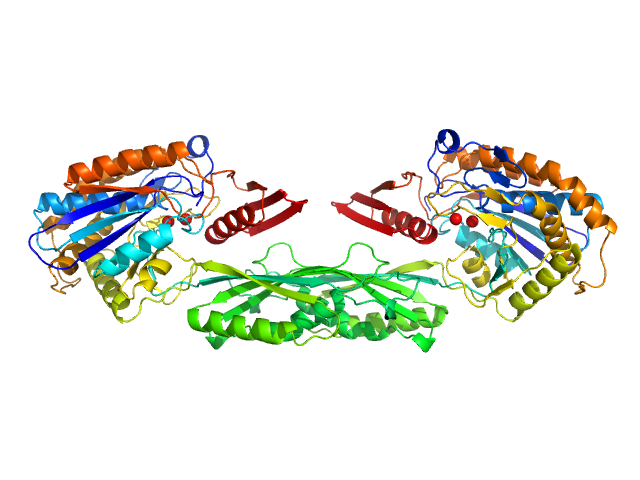
| Sample: |
Methotrexate dimer, 1 kDa
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
|
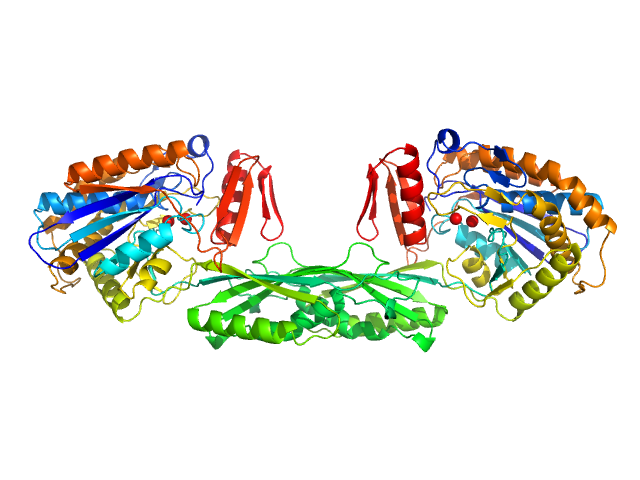
| Sample: |
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 2 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
|
|
|
|
|
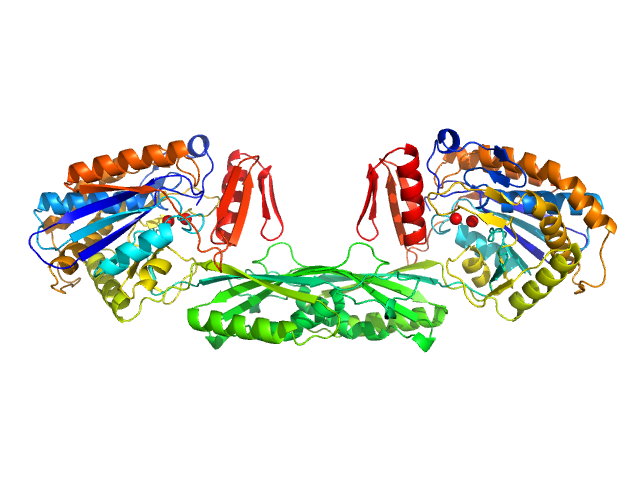
| Sample: |
Methotrexate dimer, 1 kDa
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 2 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
|
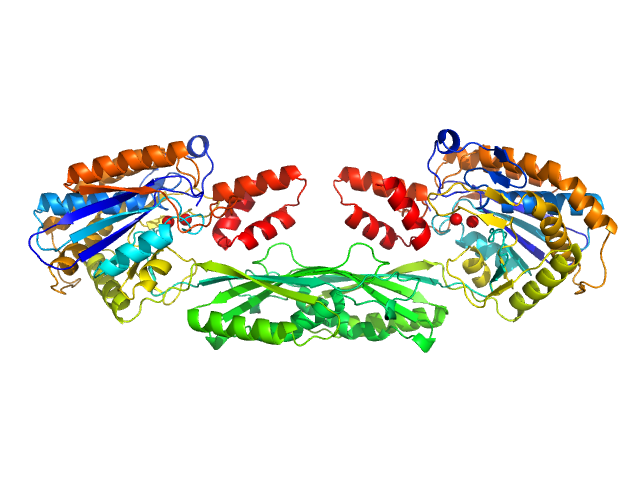
| Sample: |
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 3 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
127 |
nm3 |
|
|