|
|
|
|
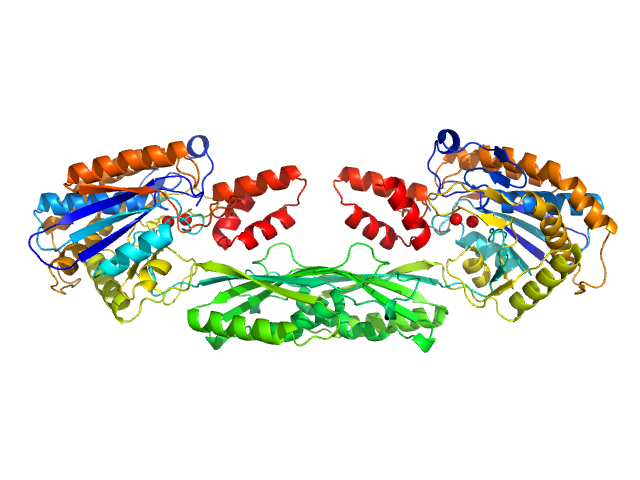
|
| Sample: |
Methotrexate dimer, 1 kDa
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 3 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
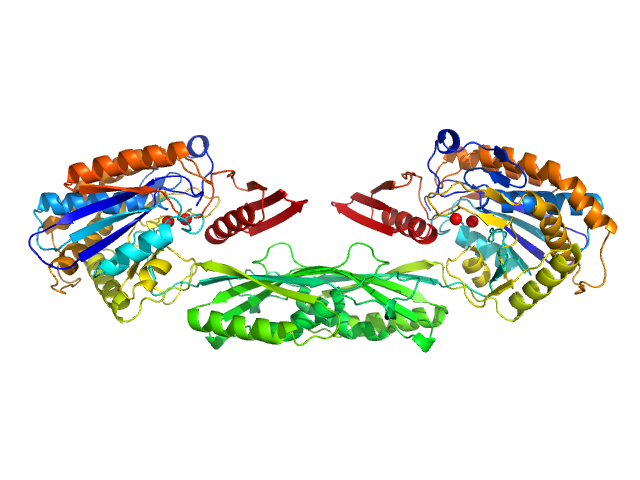
|
| Sample: |
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1 Disulfide Variant dimer, 96 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 6
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
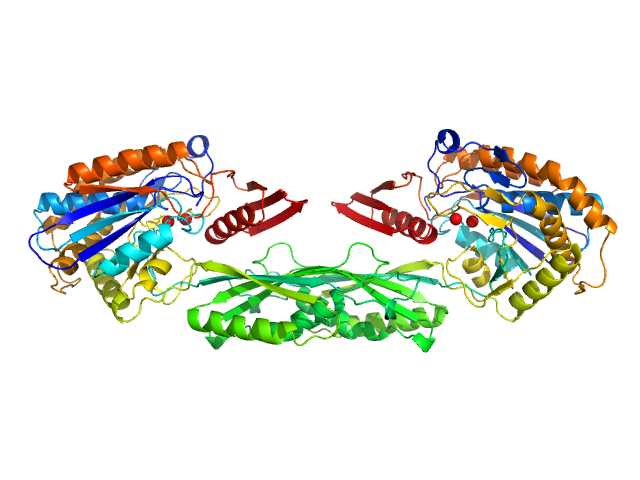
|
| Sample: |
Methotrexate dimer, 1 kDa
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1 Disulfide Variant dimer, 96 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 6
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
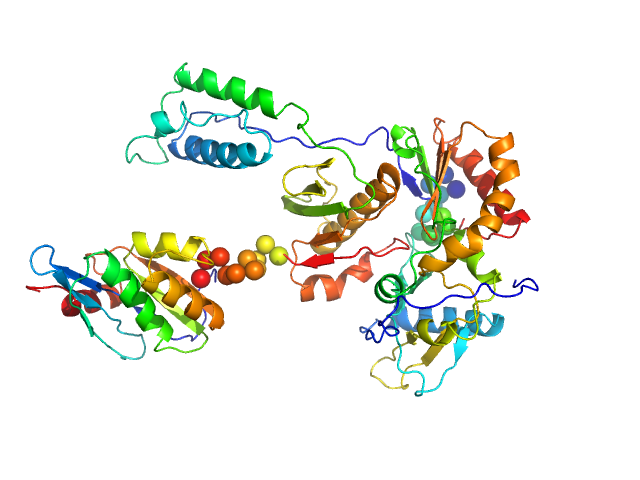
|
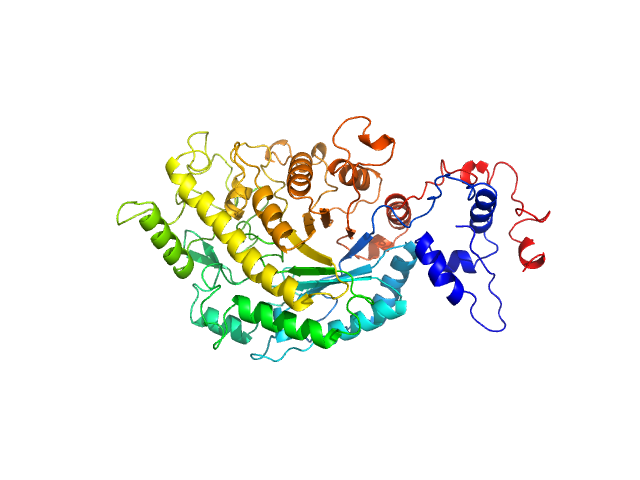
| Sample: |
Gag-Pol polyprotein (HIV-1 reverse transcriptase C38V/C280S, L289K truncated to 1 - 556) monomer, 65 kDa Human immunodeficiency virus … protein
|
| Buffer: |
50 mM Bis-Tris pH 6.8, 100 mM NaCl, 1% glycerol, 0.02% w/v NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Jun 20
|
Relative domain orientation of the L289K HIV
‐1 reverse transcriptase monomer
Protein Science 31(5) (2022)
Xi Z, Ilina T, Guerrero M, Fan L, Sluis‐Cremer N, Wang Y, Ishima R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
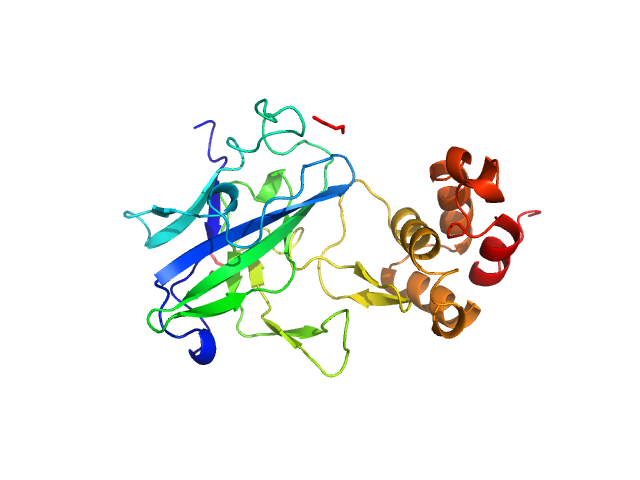
| Sample: |
Beta-amylase 1, chloroplastic monomer, 65 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM MES, 100 mM NaCl, 1 mM DTT, pH: 6.7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 22
|
The BAM7
gene in Zea mays
encodes a protein with similar structural and catalytic properties to Arabidopsis
BAM2
Acta Crystallographica Section D Structural Biology 78(5) (2022)
Ravenburg C, Riney M, Monroe J, Berndsen C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
|
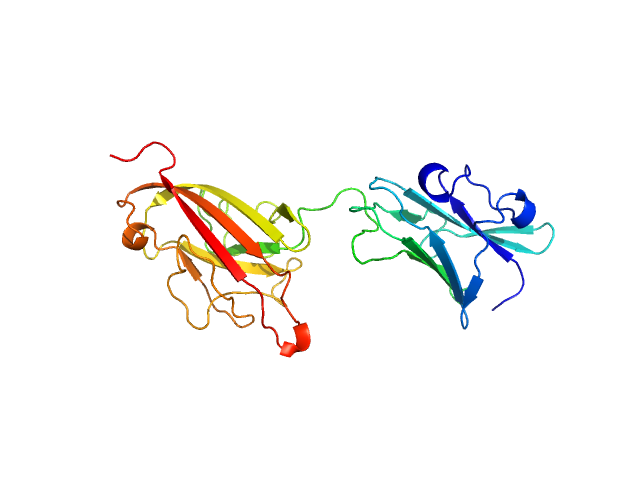
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 28
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
2.1 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
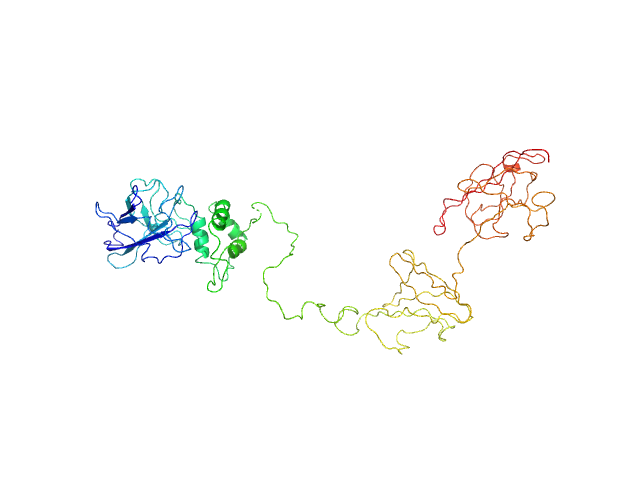
|
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 28
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
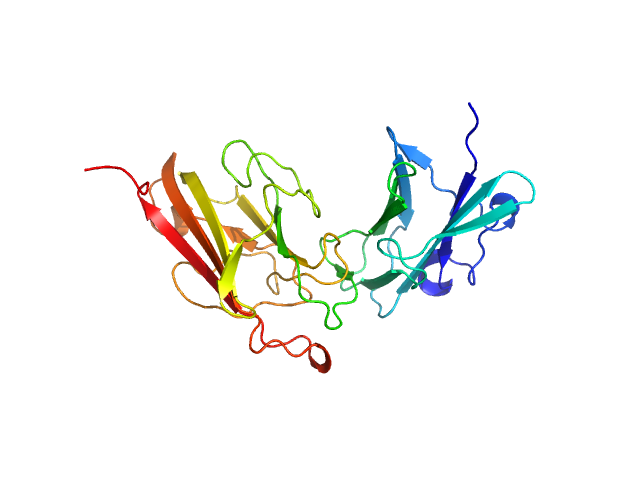
| Sample: |
Dockerin domain-containing protein monomer, 57 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 28
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
5.4 |
nm |
| Dmax |
20.3 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
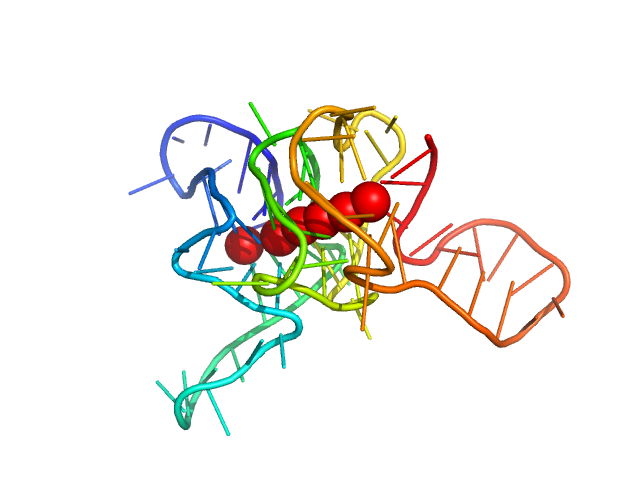
| Sample: |
C-Myc 12-tract G-quadruplex monomer Dnase I treated monomer, 22 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Sep 29
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
25 |
nm3 |
|
|