|
|
|
|

|
| Sample: |
Serotransferrin monomer, 77 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 20 mM NaHCO3, 50 mM NaCl, (APO Buffer), pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
X-ray Characterization of Conformational Changes of Human Apo- and Holo-Transferrin
International Journal of Molecular Sciences 22(24):13392 (2021)
Campos-Escamilla C, Siliqi D, Gonzalez-Ramirez L, Lopez-Sanchez C, Gavira J, Moreno A
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Serotransferrin monomer, 77 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 20 mM NaHCO3, 50 mM NaCl (APO Buffer), pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
X-ray Characterization of Conformational Changes of Human Apo- and Holo-Transferrin
International Journal of Molecular Sciences 22(24):13392 (2021)
Campos-Escamilla C, Siliqi D, Gonzalez-Ramirez L, Lopez-Sanchez C, Gavira J, Moreno A
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Serotransferrin monomer, 77 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 20 mM NaHCO3, 50 mM NaCl (Buffer APO-Tf-1), pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
X-ray Characterization of Conformational Changes of Human Apo- and Holo-Transferrin
International Journal of Molecular Sciences 22(24):13392 (2021)
Campos-Escamilla C, Siliqi D, Gonzalez-Ramirez L, Lopez-Sanchez C, Gavira J, Moreno A
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|
|
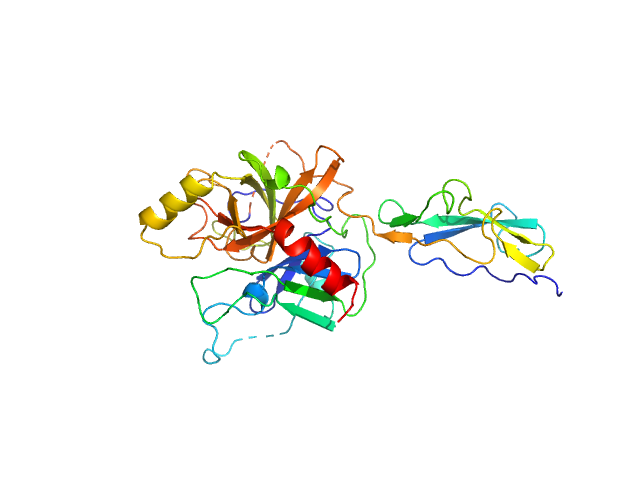
| Sample: |
Complement C1r subcomponent monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 140 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 24
|
A Structural Basis for Inhibition of the Complement Initiator Protease C1r by Lyme Disease Spirochetes.
J Immunol 207(11):2856-2867 (2021)
Garrigues RJ, Powell-Pierce AD, Hammel M, Skare JT, Garcia BL
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
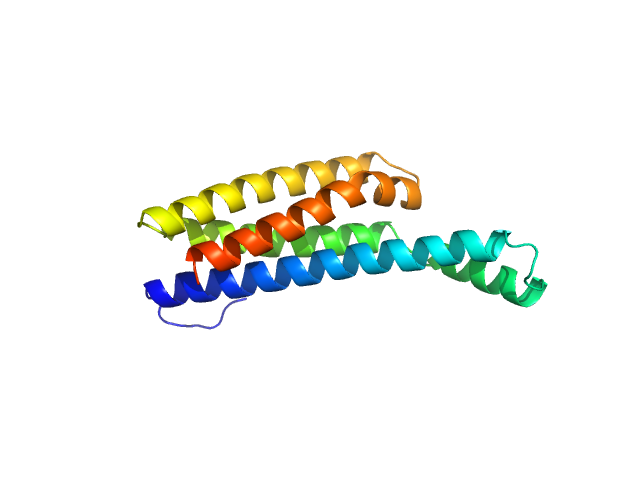
| Sample: |
Fibronectin-binding protein BBK32 monomer, 17 kDa Borreliella burgdorferi B31 protein
|
| Buffer: |
10 mM HEPES, 140 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 24
|
A Structural Basis for Inhibition of the Complement Initiator Protease C1r by Lyme Disease Spirochetes.
J Immunol 207(11):2856-2867 (2021)
Garrigues RJ, Powell-Pierce AD, Hammel M, Skare JT, Garcia BL
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
|
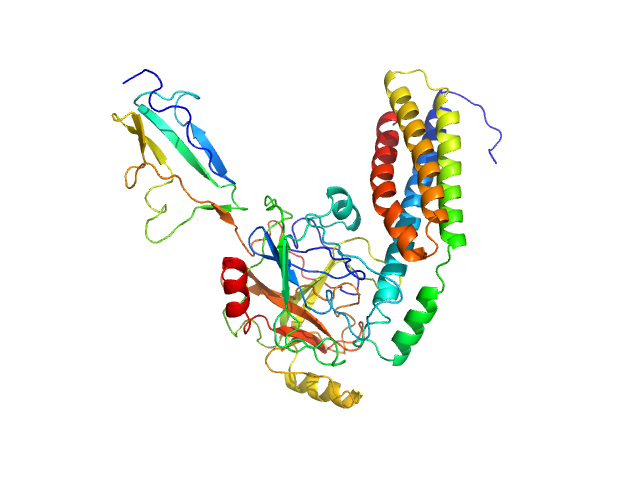
| Sample: |
Fibronectin-binding protein BBK32 monomer, 17 kDa Borrelia burgdorferi (strain … protein
Complement C1r subcomponent monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 140 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 24
|
A Structural Basis for Inhibition of the Complement Initiator Protease C1r by Lyme Disease Spirochetes.
J Immunol 207(11):2856-2867 (2021)
Garrigues RJ, Powell-Pierce AD, Hammel M, Skare JT, Garcia BL
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
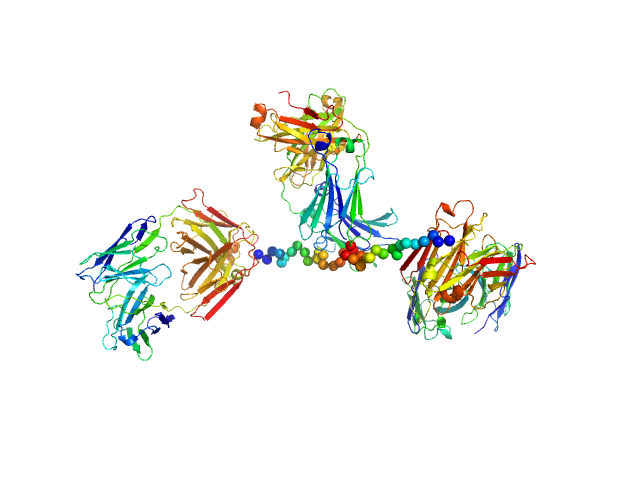
|
| Sample: |
Anti-prion protein monoclonal IgG2a 6D11 monomer, 145 kDa Mus musculus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies.
Sci Adv 7(48):eabj1826 (2021)
Linsenmeier L, Mohammadi B, Shafiq M, Frontzek K, Bär J, Shrivastava AN, Damme M, Song F, Schwarz A, Da Vela S, Massignan T, Jung S, Correia A, Schmitz M, Puig B, Hornemann S, Zerr I, Tatzelt J, Biasini E, Saftig P, Schweizer M, Svergun D, Amin L, Mazzola F, Varani L, Thapa S, Gilch S, Schätzl H, Harris DA, Triller A, Mikhaylova M, Aguzzi A, Altmeppen HC, Glatzel M
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
|
|
|
|
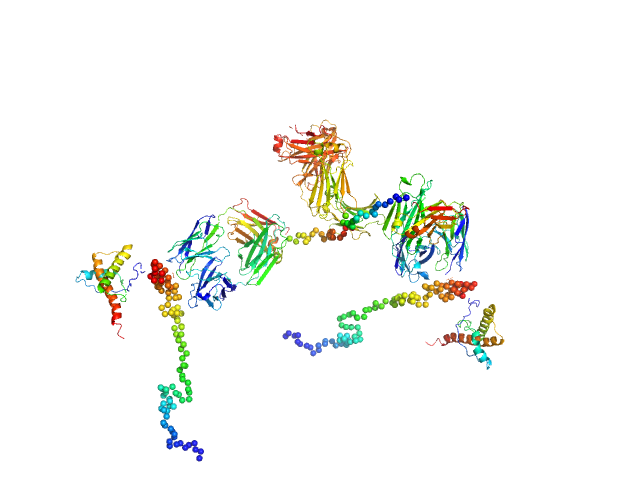
|
| Sample: |
Major prion protein monomer, 23 kDa Mus musculus protein
Anti-prion protein monoclonal IgG2a 6D11 monomer, 145 kDa Mus musculus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies.
Sci Adv 7(48):eabj1826 (2021)
Linsenmeier L, Mohammadi B, Shafiq M, Frontzek K, Bär J, Shrivastava AN, Damme M, Song F, Schwarz A, Da Vela S, Massignan T, Jung S, Correia A, Schmitz M, Puig B, Hornemann S, Zerr I, Tatzelt J, Biasini E, Saftig P, Schweizer M, Svergun D, Amin L, Mazzola F, Varani L, Thapa S, Gilch S, Schätzl H, Harris DA, Triller A, Mikhaylova M, Aguzzi A, Altmeppen HC, Glatzel M
|
| RgGuinier |
8.1 |
nm |
| Dmax |
24.8 |
nm |
| VolumePorod |
710 |
nm3 |
|
|
|
|
|
|
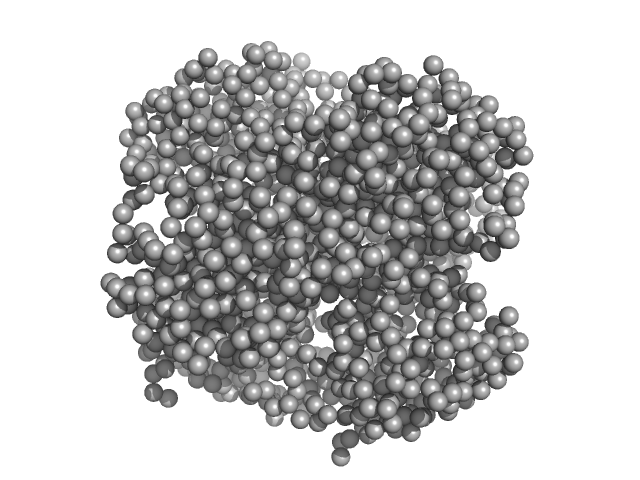
|
| Sample: |
HpcH/HpaI aldolase hexamer, 165 kDa Rhizorhabdus wittichii RW1 protein
|
| Buffer: |
20 mM HEPES,, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 23
|
EFAMIX
, a tool to decompose inline chromatography SAXS
data from partially overlapping components
Protein Science (2021)
Konarev P, Graewert M, Jeffries C, Fukuda M, Cheremnykh T, Volkov V, Svergun D
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
233 |
nm3 |
|
|
|
|
|
|
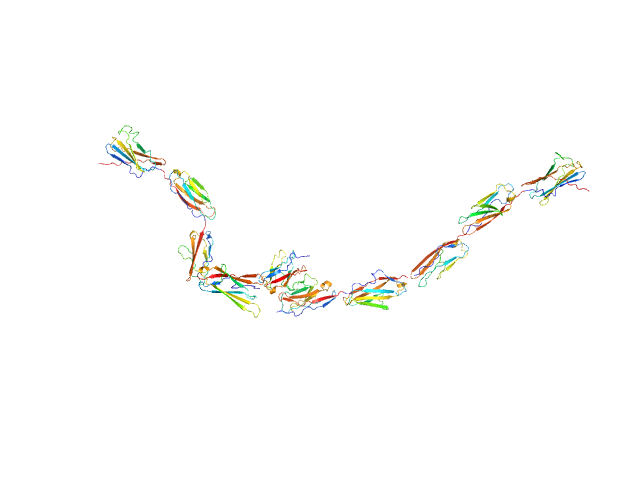
|
| Sample: |
Kin of IRRE-like protein 3 dimer, 106 kDa Mus musculus protein
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 3
|
Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep 37(5):109940 (2021)
Wang J, Vaddadi N, Pak JS, Park Y, Quilez S, Roman CA, Dumontier E, Thornton JW, Cloutier JF, Özkan E
|
| RgGuinier |
9.3 |
nm |
| Dmax |
34.5 |
nm |
|
|