|
|
|
|
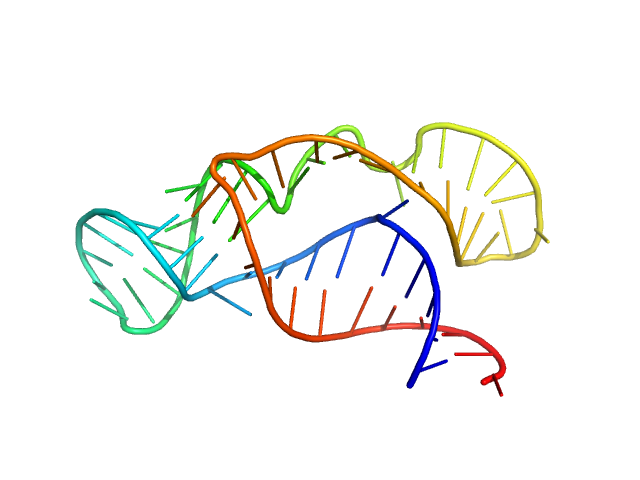
|
| Sample: |
Neurospora Varkud Satellite ribozyme junction II-III-VI monomer, 20 kDa Neurospora crassa RNA
|
| Buffer: |
50 mM MES, 50 mM KCl, 5 mM MgCl2,, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2014 Oct 29
|
An integrative NMR-SAXS approach for structural determination of large RNAs defines the substrate-free state of a trans
-cleaving Neurospora
Varkud Satellite ribozyme
Nucleic Acids Research (2021)
Dagenais P, Desjardins G, Legault P
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
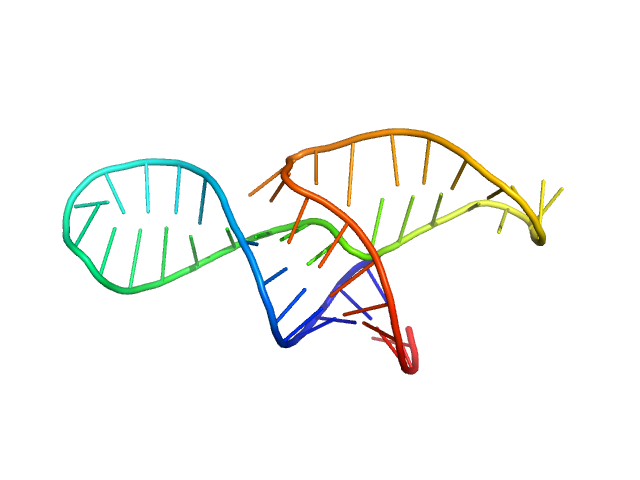
|
| Sample: |
Neurospora Varkud Satellite ribozyme junction III-IV-V monomer, 15 kDa Neurospora crassa RNA
|
| Buffer: |
50 mM MES, 50 mM KCl, 5 mM MgCl2,, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2014 Oct 29
|
An integrative NMR-SAXS approach for structural determination of large RNAs defines the substrate-free state of a trans
-cleaving Neurospora
Varkud Satellite ribozyme
Nucleic Acids Research (2021)
Dagenais P, Desjardins G, Legault P
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
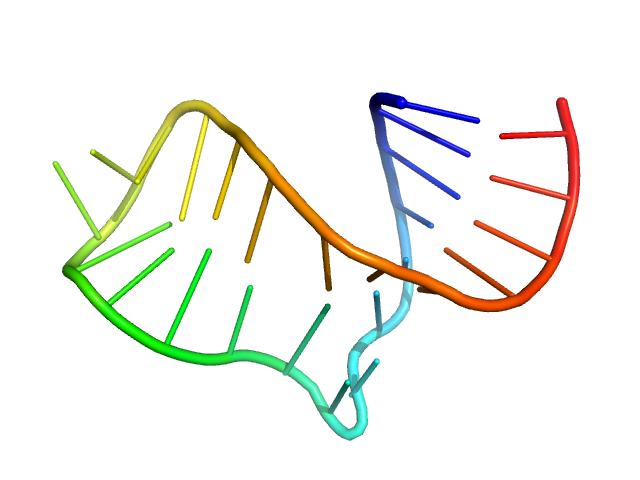
|
| Sample: |
Neurospora Varkud Satellite ribozyme stem-loop VI monomer, 8 kDa Neurospora crassa RNA
|
| Buffer: |
50 mM MES, 50 mM KCl, 5 mM MgCl2,, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Jul 7
|
An integrative NMR-SAXS approach for structural determination of large RNAs defines the substrate-free state of a trans
-cleaving Neurospora
Varkud Satellite ribozyme
Nucleic Acids Research (2021)
Dagenais P, Desjardins G, Legault P
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|
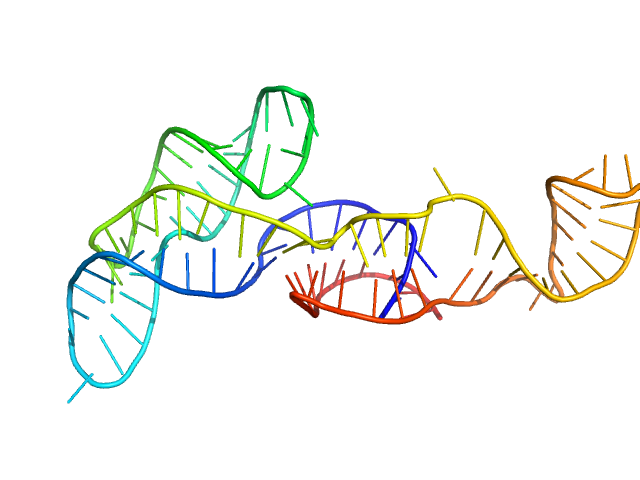
|
| Sample: |
Neurospora Varkud Satellite minimal trans ribozyme monomer, 33 kDa Neurospora crassa RNA
|
| Buffer: |
50 mM MES, 50 mM KCl, 5 mM MgCl2,, pH: 6.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2019 Sep 19
|
An integrative NMR-SAXS approach for structural determination of large RNAs defines the substrate-free state of a trans
-cleaving Neurospora
Varkud Satellite ribozyme
Nucleic Acids Research (2021)
Dagenais P, Desjardins G, Legault P
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 8
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
7.6 |
nm |
| Dmax |
41.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Jan 25
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
8.9 |
nm |
| Dmax |
39.0 |
nm |
|
|
|
|
|
|
|
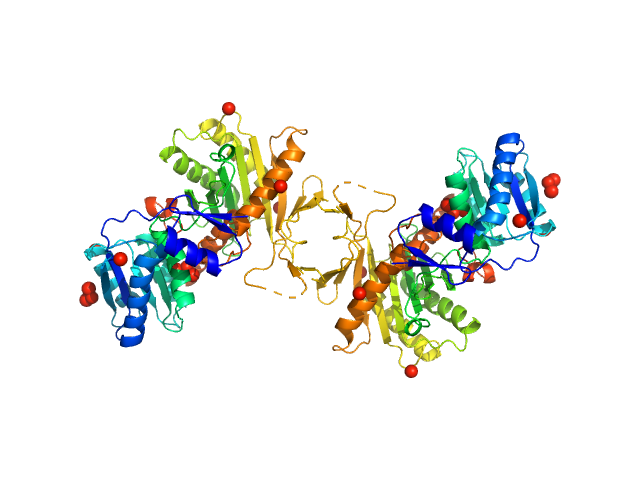
| Sample: |
Piwi protein AF_1318 dimer, 98 kDa Archaeoglobus fulgidus protein
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 500 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
Apo Archaeoglobus fulgidus Argonaute protein dimerises in solution
Elena Manakova
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
174 |
nm3 |
|
|
|
|
|
|
|
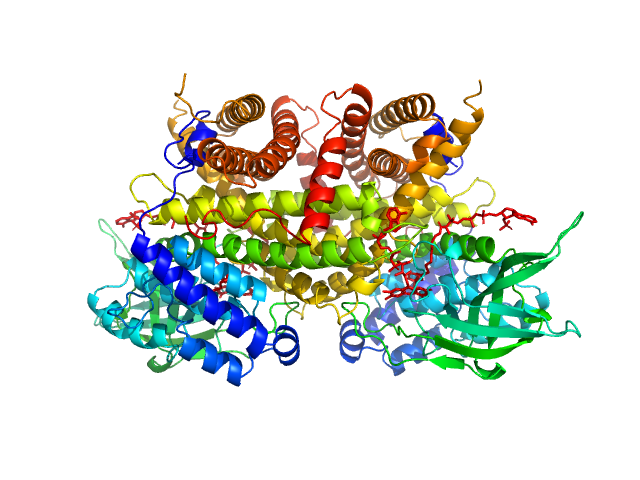
| Sample: |
Complex I assembly factor ACAD9, mitochondrial dimer, 130 kDa Homo sapiens protein
|
| Buffer: |
25mM Tris-HCl, 150mM NaCl, 0.1mM DTT, and 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 2
|
Molecular mechanism of interactions between ACAD9 and binding partners in mitochondrial respiratory complex I assembly
iScience 24(10):103153 (2021)
Xia C, Lou B, Fu Z, Mohsen A, Shen A, Vockley J, Kim J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
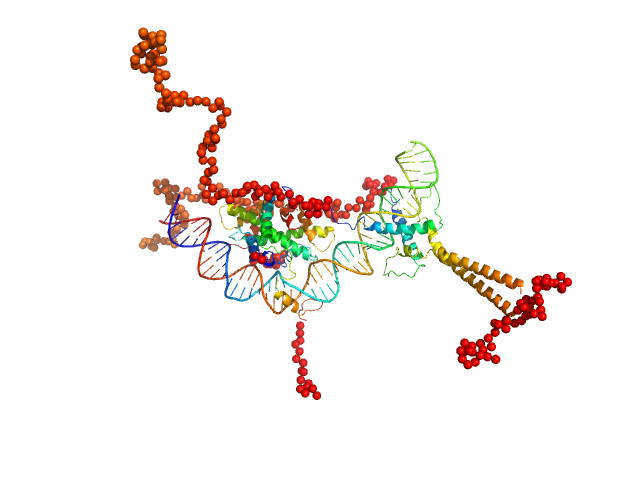
|
| Sample: |
Upstream stimulatory factor 1 dimer, 50 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 48bp monomer, 30 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Upstream stimulatory factor 1 dimer, 50 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 50bp monomer, 31 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
165 |
nm3 |
|
|