|
|
|
|
|
| Sample: |
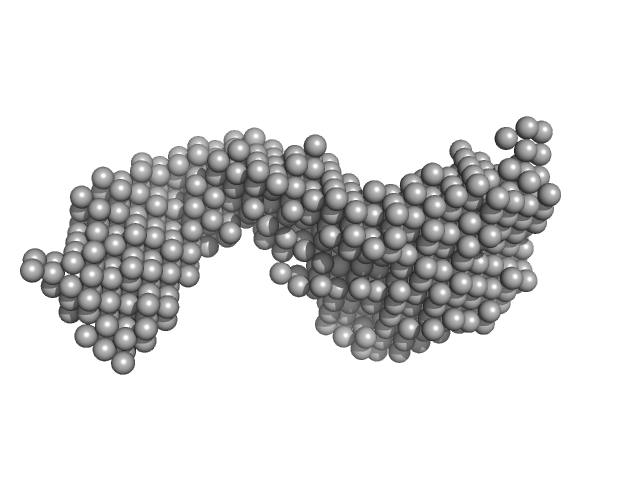
Protein max (Isoform 2, short, 13-21: missing) dimer, 39 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 48bp monomer, 30 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Dec 10
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
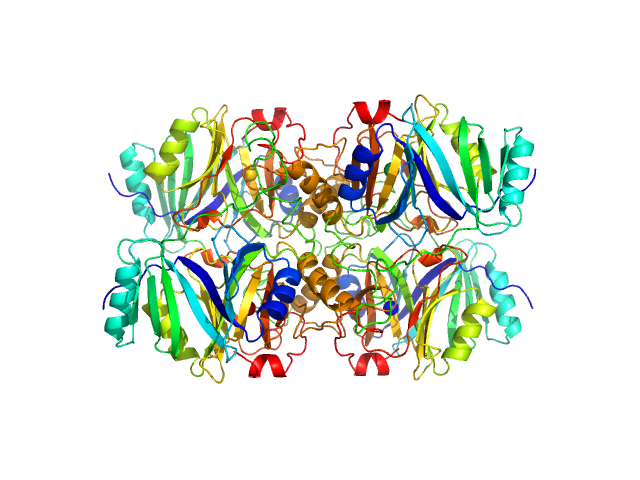
Metapyrocatechase tetramer, 139 kDa Novosphingobium sp. AAP93 protein
|
| Buffer: |
20 mM Tris-HCl 250 mM NaCl, pH 8.0, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Dec 20
|
Shape-function of a novel metapyrocatechase, RW4-MPC: Metagenomics to SAXS data based insight into deciphering regulators of function.
Int J Biol Macromol 188:1012-1024 (2021)
Vasudeva G, Sidhu C, Kalidas N, Ashish, Pinnaka AK
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
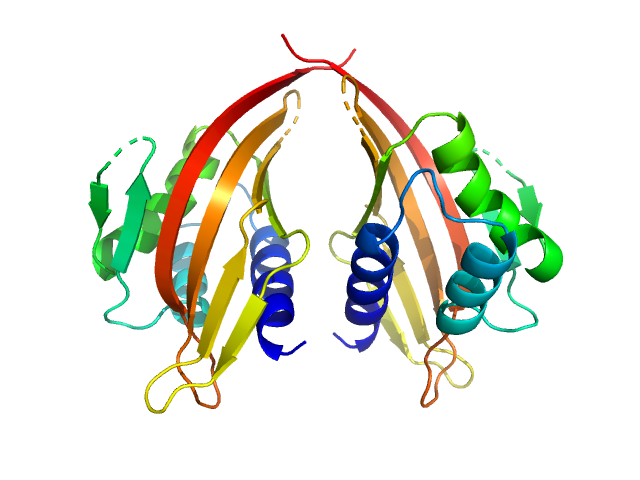
LIM domain-binding protein 1 dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Self association of LIM domain binding protein 1, LDB1
Jacqueline Matthews
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
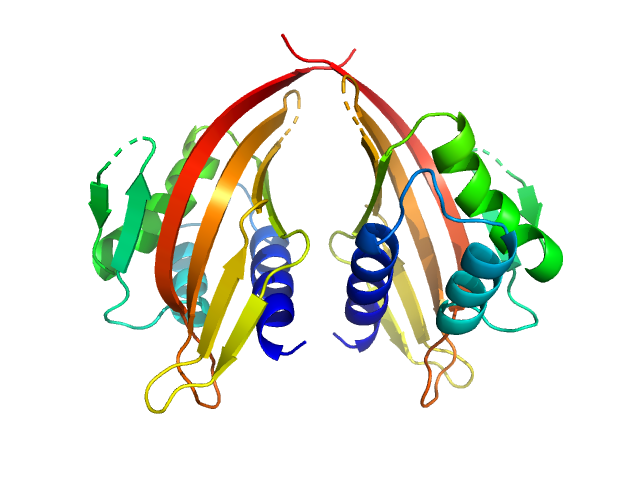
| Sample: |
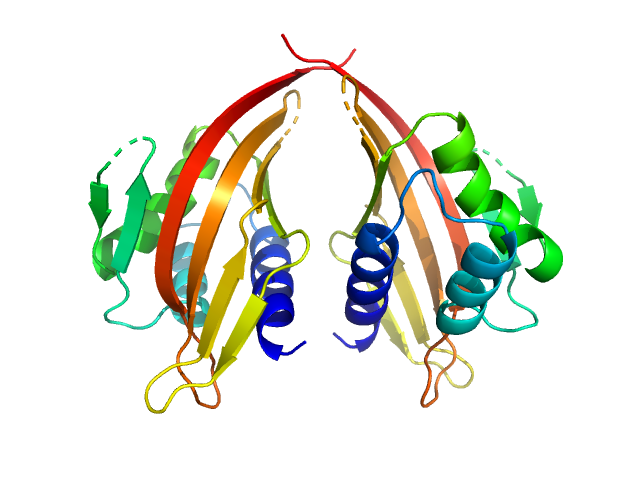
LIM domain-binding protein 1, L87E dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Self association of LIM domain binding protein 1, LDB1
Jacqueline Matthews
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
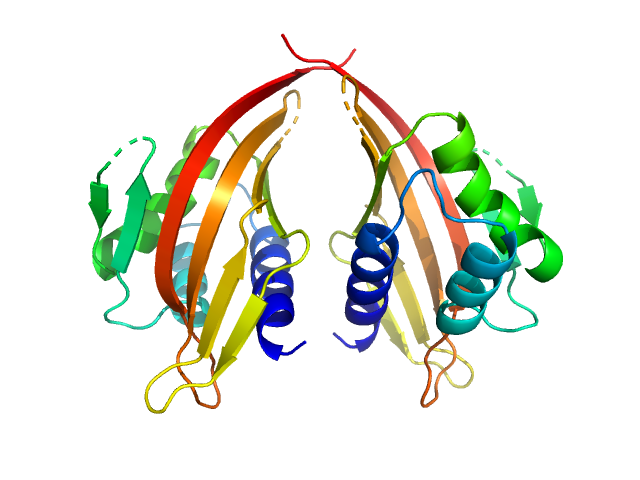
| Sample: |
LIM domain-binding protein 1, L87K dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Self association of LIM domain binding protein 1, LDB1
Jacqueline Matthews
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
LIM domain-binding protein 1, L87Q dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Self association of LIM domain binding protein 1, LDB1
Jacqueline Matthews
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
|
|
|
|
|
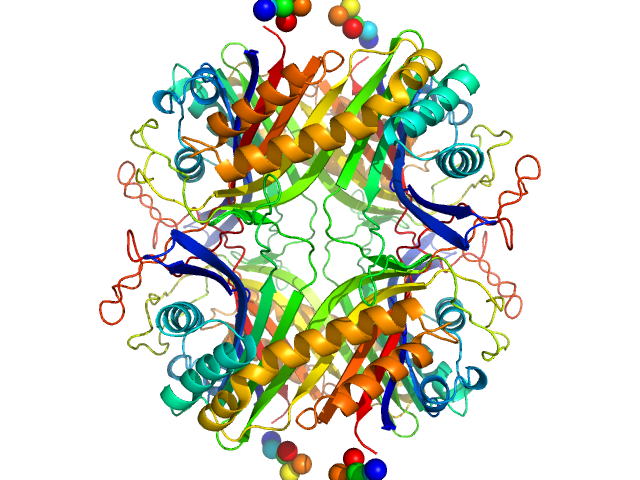
| Sample: |
Urate Oxidase (Uricase) from Aspergillus flavus tetramer, 137 kDa Aspergillus flavus protein
|
| Buffer: |
20 mM Tris. 150 mM NaCl, 1 mM EDTA, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2019 Jul 1
|
Urate Oxidase (Uricase) from Aspergillus flavus
Tsutomu Matsui
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|
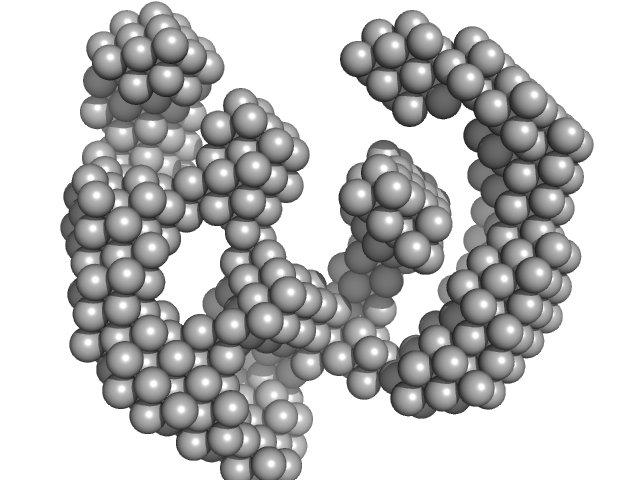
|
| Sample: |
ATP-dependent DNA helicase UvrD1 monomer, 85 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
10 mM Tris-Cl, 100 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 5% Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2020 Feb 28
|
Structural and biochemical characterization of RNA polymerase core and UvrD complex: a key component in transcription coupled DNA repair
Ravishankar Ramachandran
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
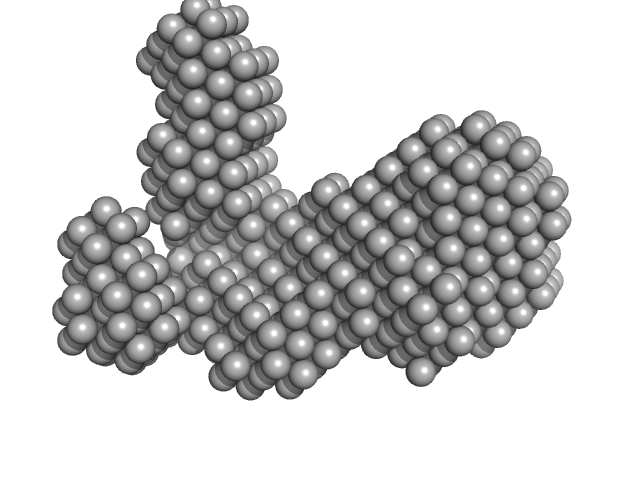
|
| Sample: |
ATP-dependent DNA helicase UvrD1 monomer, 85 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
10 mM Tris-Cl, 100 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 5% Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2020 Feb 28
|
Structural and biochemical characterization of RNA polymerase core and UvrD complex: a key component in transcription coupled DNA repair
Ravishankar Ramachandran
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
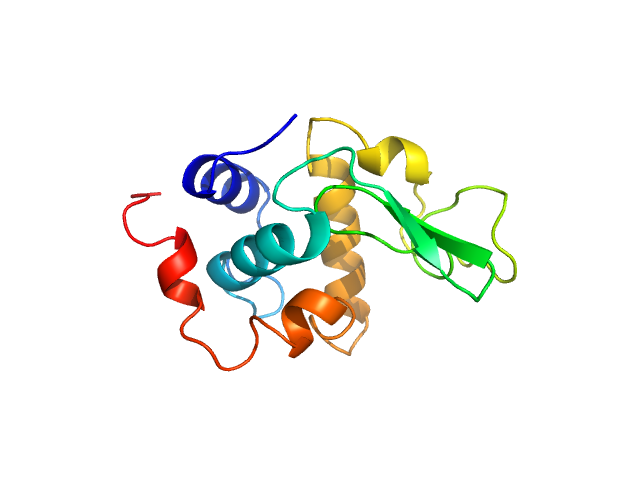
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.2 |
nm |
|
|