|
|
|
|
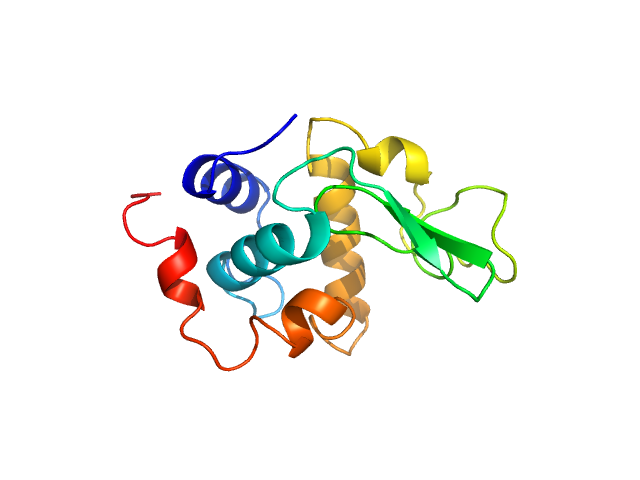
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
|
|
|
|
|
|
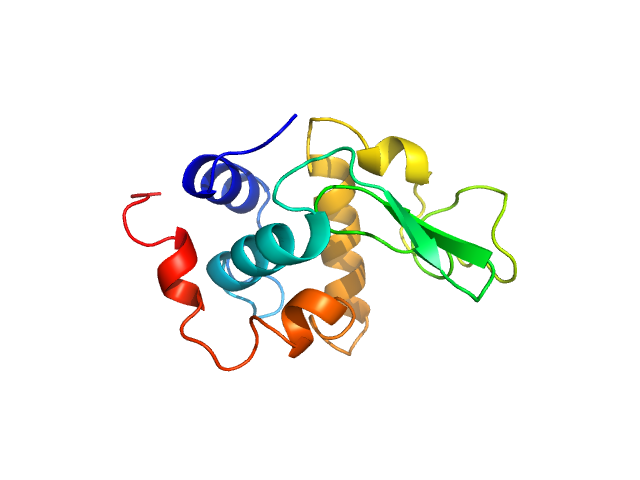
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.5 |
nm |
|
|
|
|
|
|
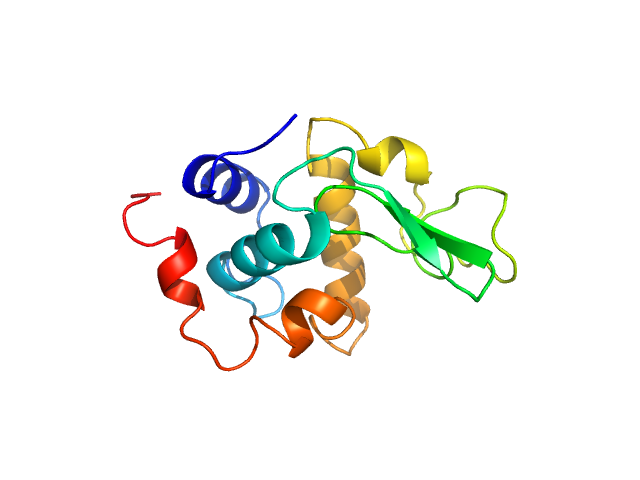
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
5.0 |
nm |
|
|
|
|
|
|
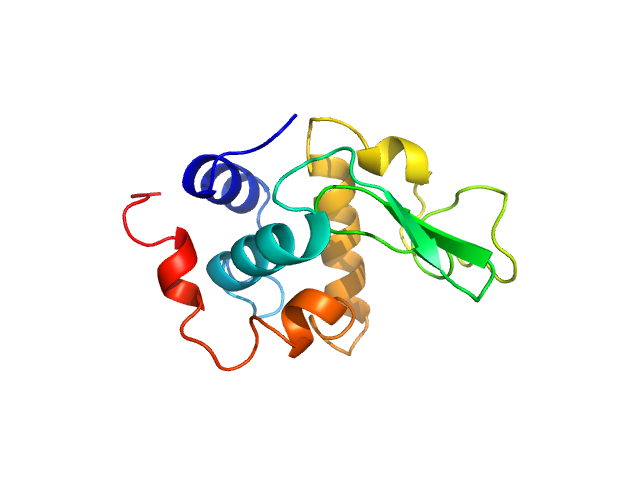
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.6 |
nm |
|
|
|
|
|
|

|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.2 |
nm |
|
|
|
|
|
|

|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.2 |
nm |
|
|
|
|
|
|

|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.2 |
nm |
|
|
|
|
|
|
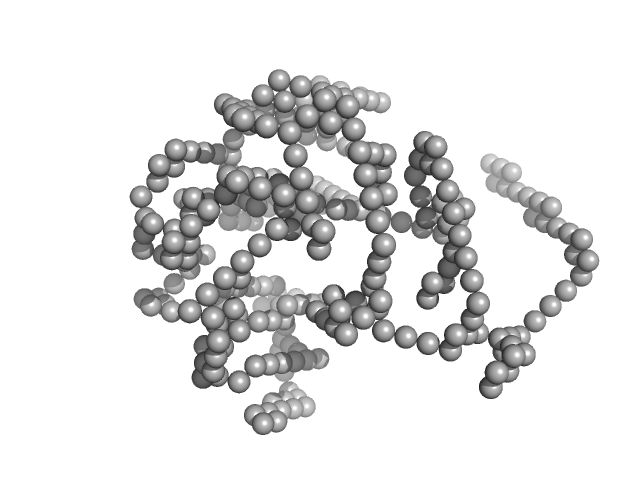
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.2 |
nm |
|
|
|
|
|
|
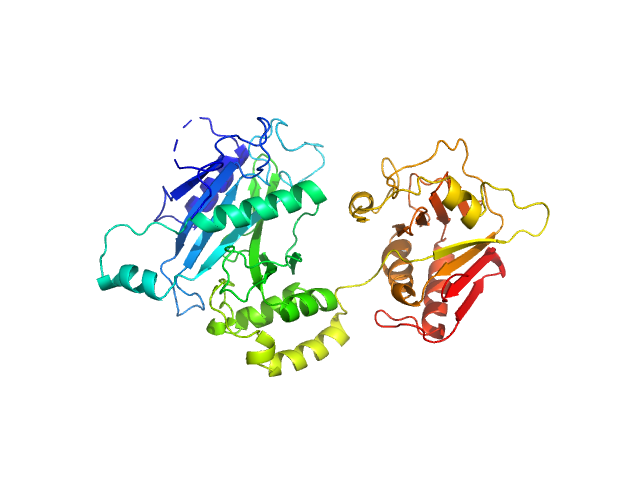
|
| Sample: |
Suppressor of fused homolog monomer, 53 kDa Drosophila melanogaster protein
|
| Buffer: |
50 mM bis-TRIS pH 5.5, 200 mM NaCl, 10% glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 May 7
|
Suppressor of Fused
Valerie Biou
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
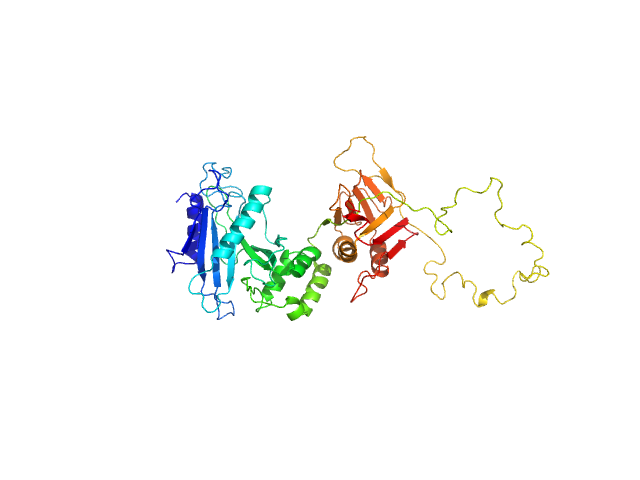
|
| Sample: |
Suppressor of fused homolog monomer, 51 kDa Homo sapiens protein
|
| Buffer: |
50 mM bis-TRIS pH 5.5, 200 mM NaCl, 10% glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Jun 30
|
Suppressor of Fused
Valerie Biou
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.0 |
nm |
|
|