|
|
|
|
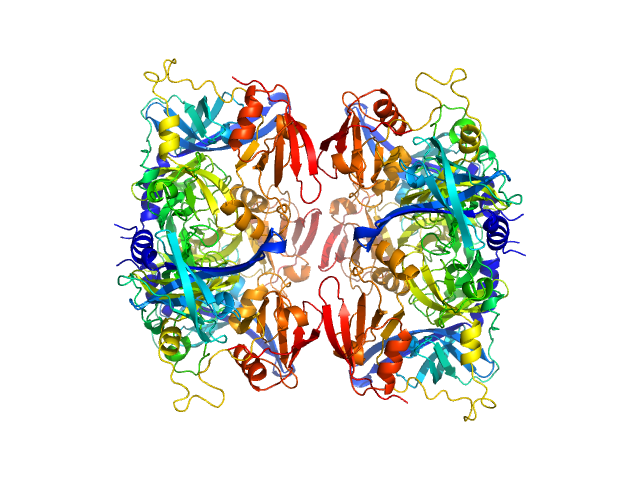
|
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
250 |
nm3 |
|
|
|
|
|
|
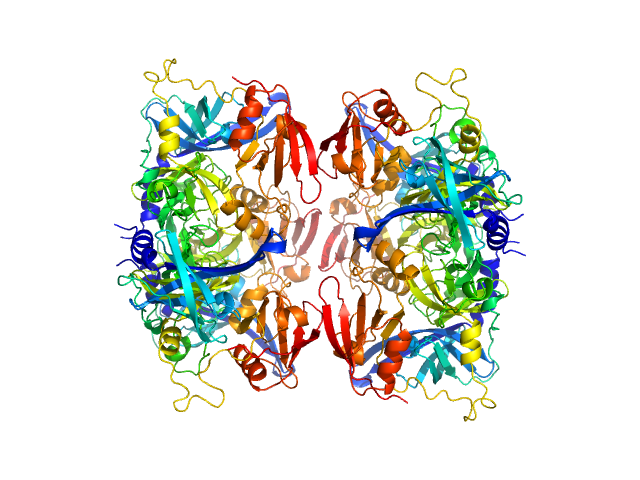
|
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
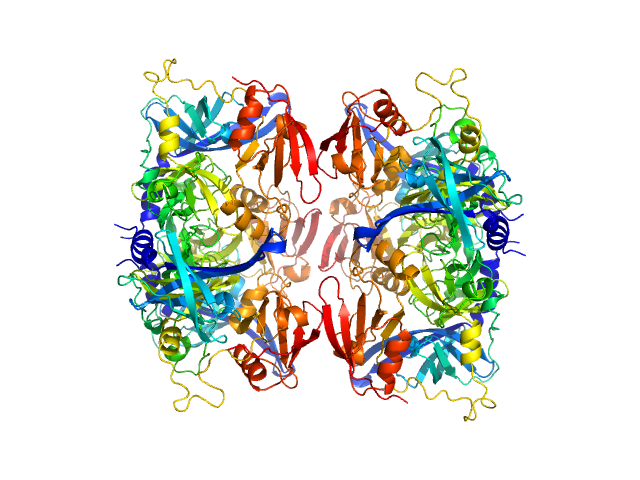
|
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
203 |
nm3 |
|
|
|
|
|
|
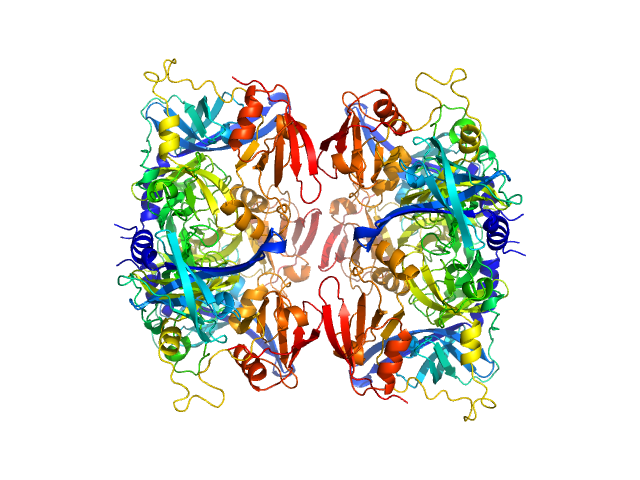
|
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|

|
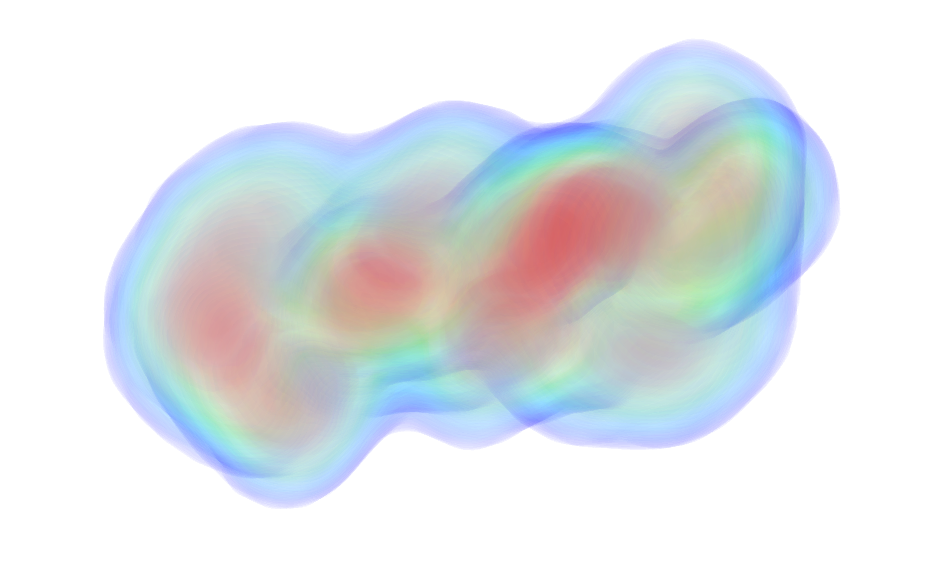
| Sample: |
DNA protection during starvation protein dodecamer, 225 kDa Escherichia coli protein
|
| Buffer: |
20мМ Tris-HCl, 100 мМ NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 19
|
Spatial Organization of Dps and DNA-Dps Complexes.
J Mol Biol :166930 (2021)
Dubrovin EV, Dadinova LA, Petoukhov MV, Yu Soshinskaya E, Mozhaev AA, Klinov DV, Schäffer TE, Shtykova EV, Batishchev OV
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Bifunctional hemolysin/adenylate cyclase monomer, 3 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 4 mM CaCl₂, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Sep 24
|
A High‐Affinity Calmodulin‐Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells
Advanced Science :2003630 (2021)
Voegele A, Sadi M, O'Brien D, Gehan P, Raoux‐Barbot D, Davi M, Hoos S, Brûlé S, Raynal B, Weber P, Mechaly A, Haouz A, Rodriguez N, Vachette P, Durand D, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
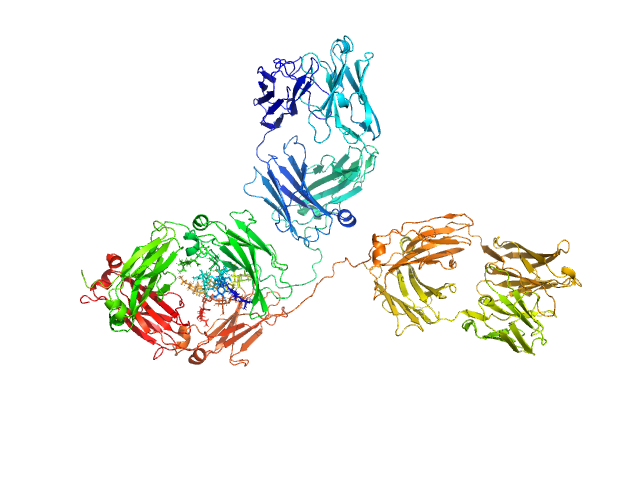
| Sample: |
Immunoglobulin G subclass 1, 148 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 20
|
Solution structure of deglycosylated human IgG1 shows the role of CH2 glycans in its conformation
Biophysical Journal (2021)
Spiteri V, Doutch J, Rambo R, Gor J, Dalby P, Perkins S
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
269 |
nm3 |
|
|
|
|
|
|
|
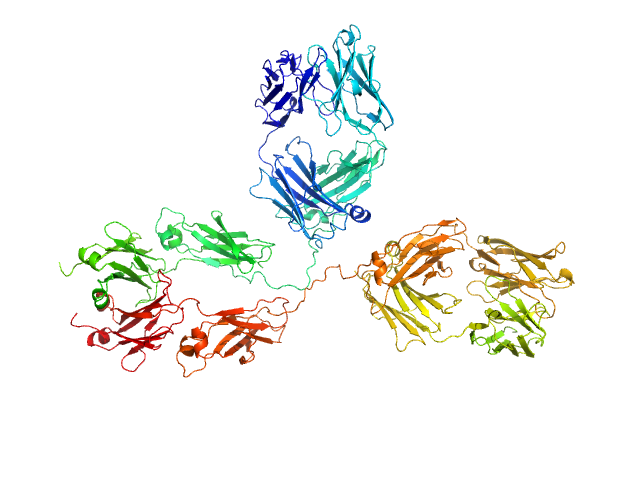
| Sample: |
Immunoglobulin G subclass 1, 148 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 20
|
Solution structure of deglycosylated human IgG1 shows the role of CH2 glycans in its conformation
Biophysical Journal (2021)
Spiteri V, Doutch J, Rambo R, Gor J, Dalby P, Perkins S
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
271 |
nm3 |
|
|
|
|
|
|
|
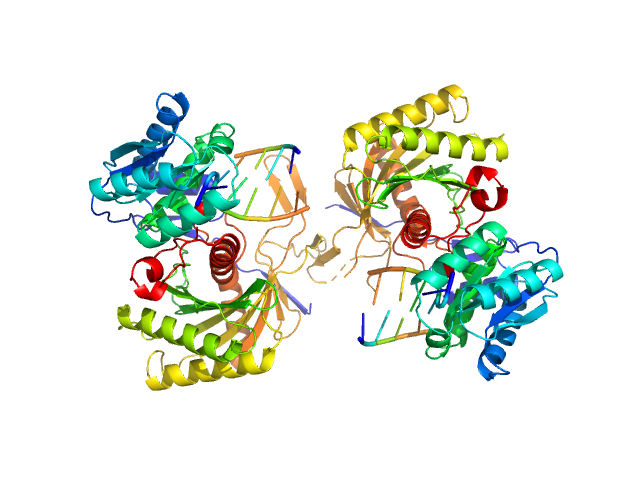
| Sample: |
Piwi protein AF_1318 dimer, 102 kDa Archaeoglobus fulgidus protein
5'-phosphorilated 14-mer DNA oligoduplex dimer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
Prokaryotic Argonaute from Archaeoglobus fulgidus interacts with DNA as a homodimer.
Sci Rep 11(1):4518 (2021)
Golovinas E, Rutkauskas D, Manakova E, Jankunec M, Silanskas A, Sasnauskas G, Zaremba M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
158 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Piwi protein AF_1318 delta (296-303) mutant monomer, 50 kDa Archaeoglobus fulgidus protein
5'-phosphorilated 14-mer DNA oligoduplex dimer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
Prokaryotic Argonaute from Archaeoglobus fulgidus interacts with DNA as a homodimer.
Sci Rep 11(1):4518 (2021)
Golovinas E, Rutkauskas D, Manakova E, Jankunec M, Silanskas A, Sasnauskas G, Zaremba M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
109 |
nm3 |
|
|