|
|
|
|
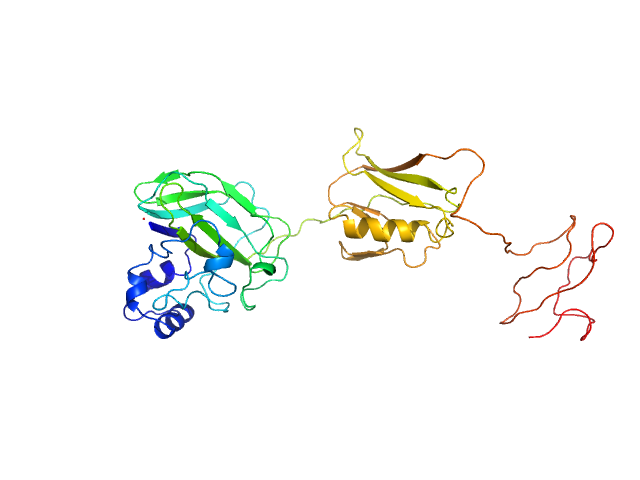
|
| Sample: |
Chitin-binding protein CbpD monomer, 39 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
15 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar with InCoatec Cu microsource, RECX, University of Oslo on 2019 Jul 19
|
The lytic polysaccharide monooxygenase CbpD promotes Pseudomonas aeruginosa virulence in systemic infection.
Nat Commun 12(1):1230 (2021)
Askarian F, Uchiyama S, Masson H, Sørensen HV, Golten O, Bunæs AC, Mekasha S, Røhr ÅK, Kommedal E, Ludviksen JA, Arntzen MØ, Schmidt B, Zurich RH, van Sorge NM, Eijsink VGH, Krengel U, Mollnes TE, Lewis NE, Nizet V, Vaaje-Kolstad G
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
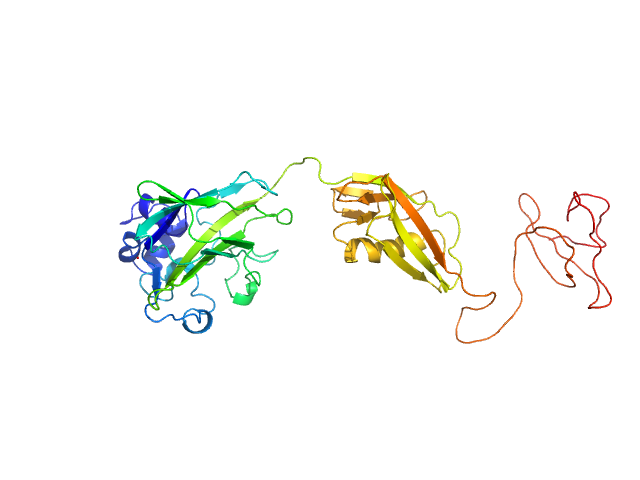
|
| Sample: |
Chitin-binding protein CbpD monomer, 39 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
15 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Oct 1
|
The lytic polysaccharide monooxygenase CbpD promotes Pseudomonas aeruginosa virulence in systemic infection.
Nat Commun 12(1):1230 (2021)
Askarian F, Uchiyama S, Masson H, Sørensen HV, Golten O, Bunæs AC, Mekasha S, Røhr ÅK, Kommedal E, Ludviksen JA, Arntzen MØ, Schmidt B, Zurich RH, van Sorge NM, Eijsink VGH, Krengel U, Mollnes TE, Lewis NE, Nizet V, Vaaje-Kolstad G
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
64131 |
nm3 |
|
|
|
|
|
|
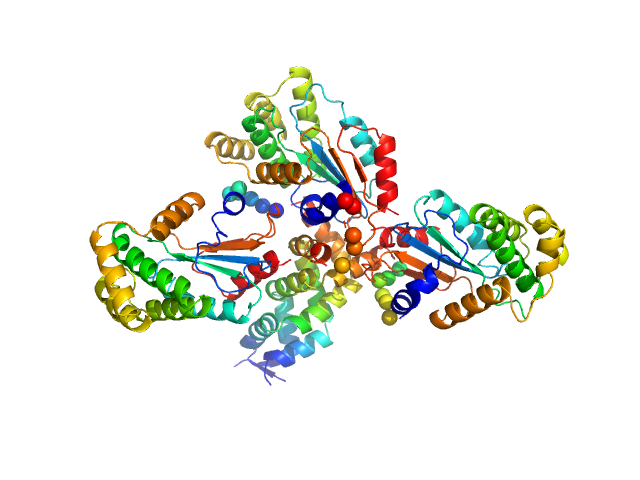
|
| Sample: |
Hypothetical exported protein trimer, 83 kDa Salmonella typhimurium (strain … protein
|
| Buffer: |
25 mM TRIS, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 14
|
Salmonella enterica BcfH is a trimeric thioredoxin-like bifunctional enzyme with both thiol oxidase and disulfide isomerase activities.
Antioxid Redox Signal (2021)
Subedi P, Paxman JJ, Wang G, Hor L, Hong Y, Verderosa AD, Whitten AE, Panjikar S, Santos-Martin CF, Martin JL, Totsika M, Heras B
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
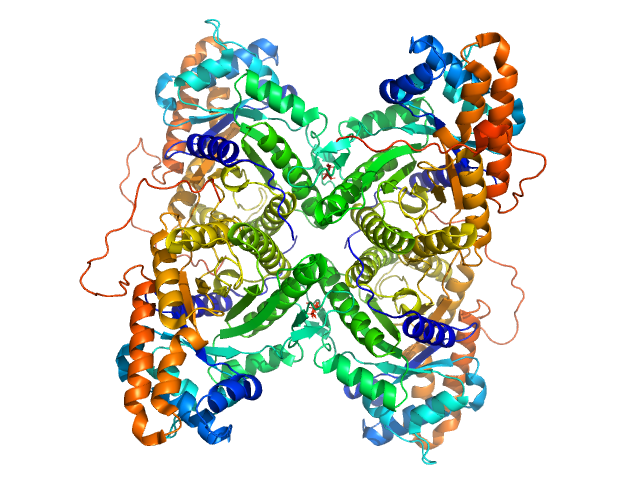
|
| Sample: |
Fructose-bisphosphate aldolase A tetramer, 157 kDa Oryctolagus cuniculus protein
|
| Buffer: |
20 mM HEPES, pH: 7 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2019 Jun 14
|
Rabbit muscle fructose-1,6-bisphosphate aldolase K229M mutant
Normand Cyr
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
|
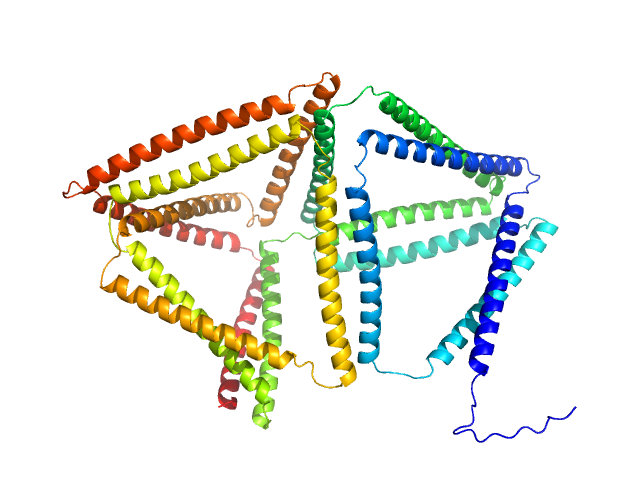
| Sample: |
BIP18SN monomer, 80 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Nov 28
|
Self-assembly and regulation of protein cages from pre-organised coiled-coil modules.
Nat Commun 12(1):939 (2021)
Lapenta F, Aupič J, Vezzoli M, Strmšek Ž, Da Vela S, Svergun DI, Carazo JM, Melero R, Jerala R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
12.4 |
nm |
|
|
|
|
|
|
|
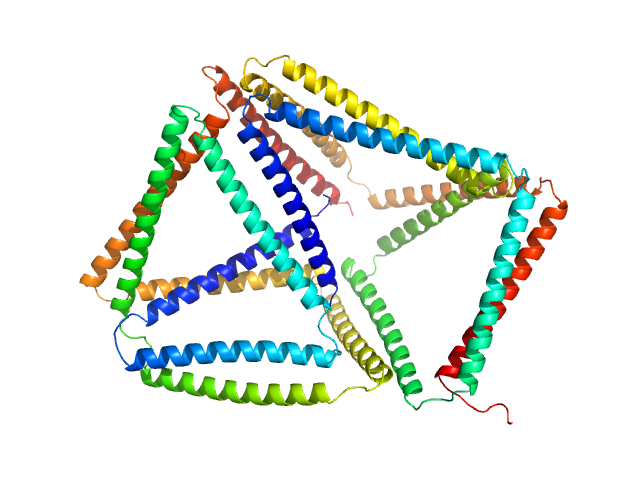
| Sample: |
SBP1(9.b) monomer, 40 kDa synthetic construct protein
SBP2(9.b) monomer, 40 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 19
|
Self-assembly and regulation of protein cages from pre-organised coiled-coil modules.
Nat Commun 12(1):939 (2021)
Lapenta F, Aupič J, Vezzoli M, Strmšek Ž, Da Vela S, Svergun DI, Carazo JM, Melero R, Jerala R
|
| RgGuinier |
4.0 |
nm |
| Dmax |
11.8 |
nm |
|
|
|
|
|
|
|
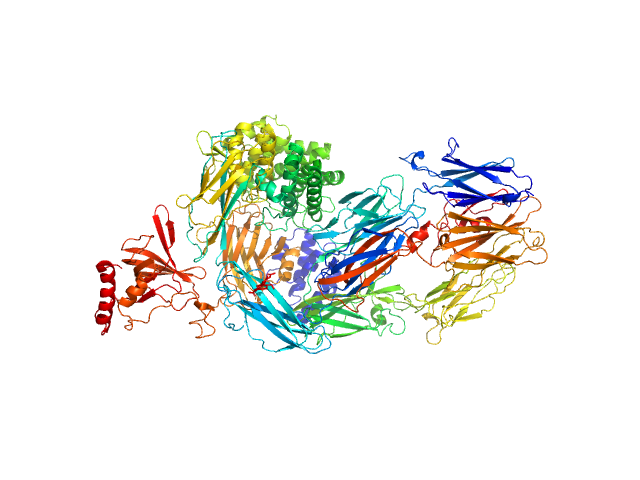
| Sample: |
Complement C5 monomer, 186 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris pH, 75mM NaCl, and 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 12
|
The allosteric modulation of Complement C5 by knob domain peptides.
Elife 10 (2021)
Macpherson A, Laabei M, Ahdash Z, Graewert MA, Birtley JR, Schulze ME, Crennell S, Robinson SA, Holmes B, Oleinikovas V, Nilsson PH, Snowden J, Ellis V, Mollnes TE, Deane CM, Svergun D, Lawson AD, van den Elsen JM
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
392 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
TET12(1.10)SN-f5(2CC) monomer, 55 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 13
|
Designed folding pathway of modular coiled-coil-based proteins.
Nat Commun 12(1):940 (2021)
Aupič J, Strmšek Ž, Lapenta F, Pahovnik D, Pisanski T, Drobnak I, Ljubetič A, Jerala R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
158 |
nm3 |
|
|
|
|
|
|

|
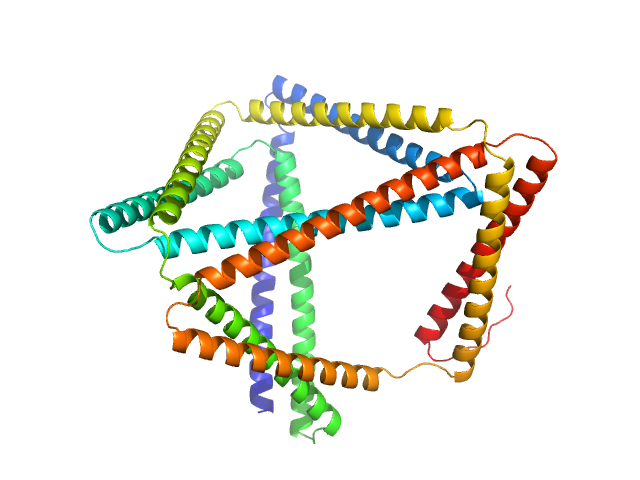
| Sample: |
TET12(1.10)SN-f5(22CC) monomer, 55 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 5
|
Designed folding pathway of modular coiled-coil-based proteins.
Nat Commun 12(1):940 (2021)
Aupič J, Strmšek Ž, Lapenta F, Pahovnik D, Pisanski T, Drobnak I, Ljubetič A, Jerala R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
TET12(1.10)SN-f5(222CC) monomer, 55 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 15
|
Designed folding pathway of modular coiled-coil-based proteins.
Nat Commun 12(1):940 (2021)
Aupič J, Strmšek Ž, Lapenta F, Pahovnik D, Pisanski T, Drobnak I, Ljubetič A, Jerala R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
170 |
nm3 |
|
|