|
|
|
|
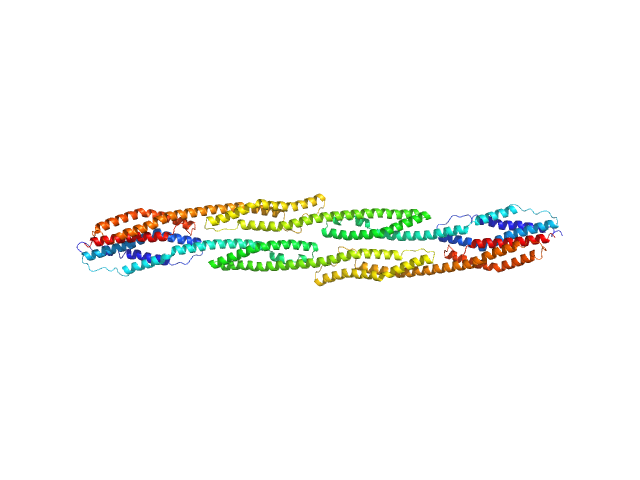
|
| Sample: |
Rod domain of α-actinin-2 dimer, 112 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 5
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
6.7 |
nm |
| Dmax |
27.2 |
nm |
| VolumePorod |
214 |
nm3 |
|
|
|
|
|
|
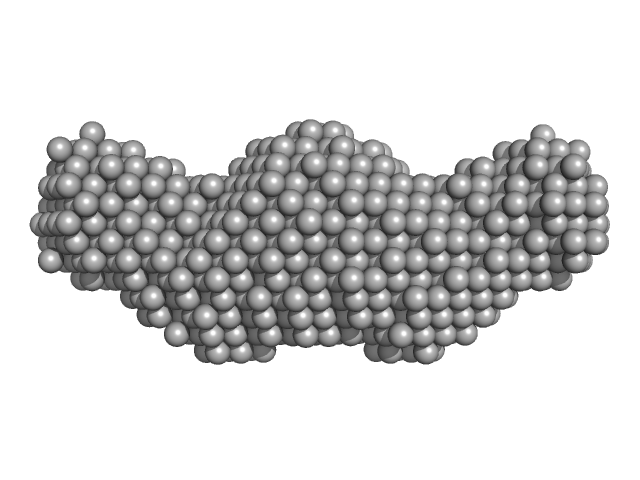
|
| Sample: |
Zinc transporter 1 dimer, 37 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris, 150mM NaCl, 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 14
|
Heterologous Expression and Biochemical Characterization of the Human Zinc Transporter 1 (ZnT1) and Its Soluble C-Terminal Domain
Frontiers in Chemistry 9 (2021)
Cotrim C, Jarrott R, Whitten A, Choudhury H, Drew D, Martin J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
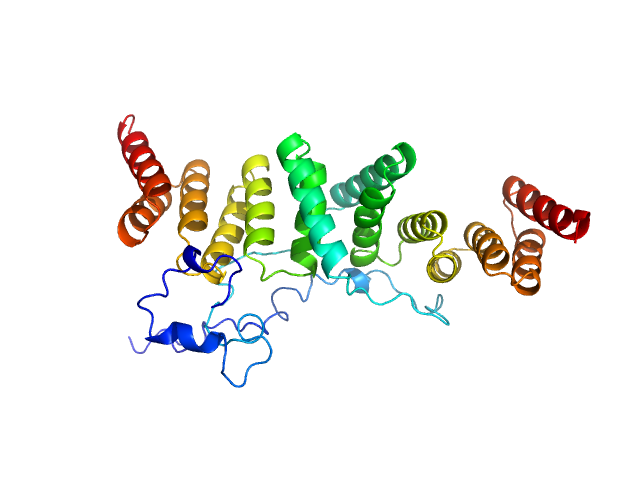
|
| Sample: |
Chaperone protein IpgC dimer, 39 kDa Shigella flexneri protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Dec 10
|
Structural Insights of Shigella Translocator IpaB and Its Chaperone IpgC in Solution
Frontiers in Cellular and Infection Microbiology 11 (2021)
Ferrari M, Charova S, Sansonetti P, Mylonas E, Gazi A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
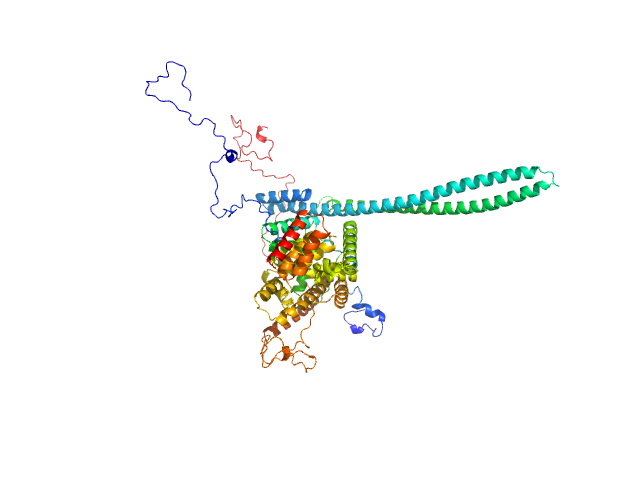
|
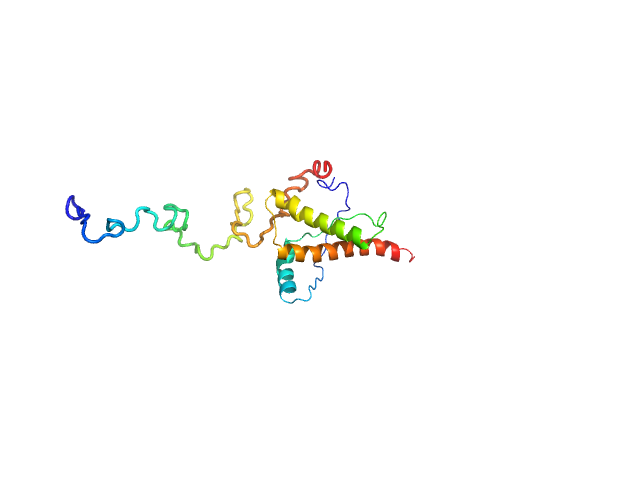
| Sample: |
T3SS Chaperone protein IpgC monomer, 20 kDa Shigella flexneri protein
Invasin IpaB monomer, 62 kDa Shigella flexneri protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Dec 10
|
Structural Insights of Shigella Translocator IpaB and Its Chaperone IpgC in Solution
Frontiers in Cellular and Infection Microbiology 11 (2021)
Ferrari M, Charova S, Sansonetti P, Mylonas E, Gazi A
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Major prion protein monomer, 23 kDa Mus musculus protein
|
| Buffer: |
10mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Characterization of murine prion protein
Stefano Da Vela
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Rationally optimised WA20 mutant N22A/H86K (ROWA) dimer dimer, 25 kDa Artificial protein (de … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Nov 30
|
Rational thermostabilisation of four-helix bundle dimeric de novo proteins.
Sci Rep 11(1):7526 (2021)
Irumagawa S, Kobayashi K, Saito Y, Miyata T, Umetsu M, Kameda T, Arai R
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Rationally optimised SUWA mutant N22E/H86K (ROSA) dimer dimer, 25 kDa Artificial protein (de … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Nov 30
|
Rational thermostabilisation of four-helix bundle dimeric de novo proteins.
Sci Rep 11(1):7526 (2021)
Irumagawa S, Kobayashi K, Saito Y, Miyata T, Umetsu M, Kameda T, Arai R
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
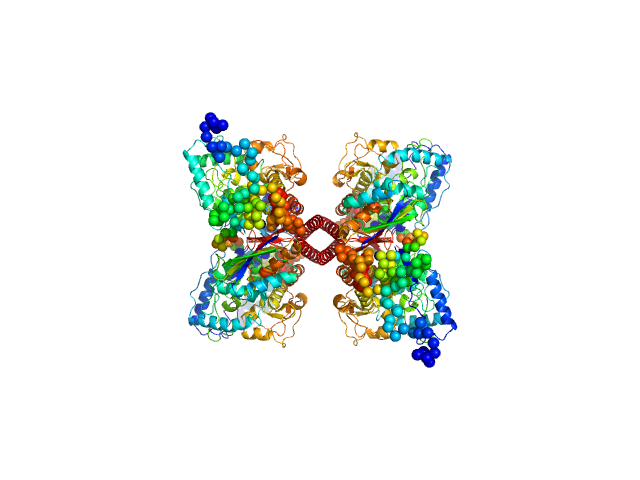
|
| Sample: |
Phenylalanine-4-hydroxylase tetramer, 223 kDa Homo sapiens protein
|
| Buffer: |
0.02 mM Hepes, 0.2 M NaCl,, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Feb 25
|
Modulation of Human Phenylalanine Hydroxylase by 3-Hydroxyquinolin-2(1H)-One Derivatives
Biomolecules 11(3):462 (2021)
Lopes R, Tomé C, Russo R, Paterna R, Leandro J, Candeias N, Gonçalves L, Teixeira M, Sousa P, Guedes R, Vicente J, Gois P, Leandro P
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
336 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Phenylalanine-4-hydroxylase tetramer, 223 kDa Homo sapiens protein
|
| Buffer: |
0.02 mM Hepes, 0.2 M NaCl,, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Feb 25
|
Modulation of Human Phenylalanine Hydroxylase by 3-Hydroxyquinolin-2(1H)-One Derivatives
Biomolecules 11(3):462 (2021)
Lopes R, Tomé C, Russo R, Paterna R, Leandro J, Candeias N, Gonçalves L, Teixeira M, Sousa P, Guedes R, Vicente J, Gois P, Leandro P
|
| RgGuinier |
4.0 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
367 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA protection during starvation protein dodecamer, 225 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 26
|
Spatial Organization of Dps and DNA-Dps Complexes.
J Mol Biol :166930 (2021)
Dubrovin EV, Dadinova LA, Petoukhov MV, Yu Soshinskaya E, Mozhaev AA, Klinov DV, Schäffer TE, Shtykova EV, Batishchev OV
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
314 |
nm3 |
|
|