|
|
|
|
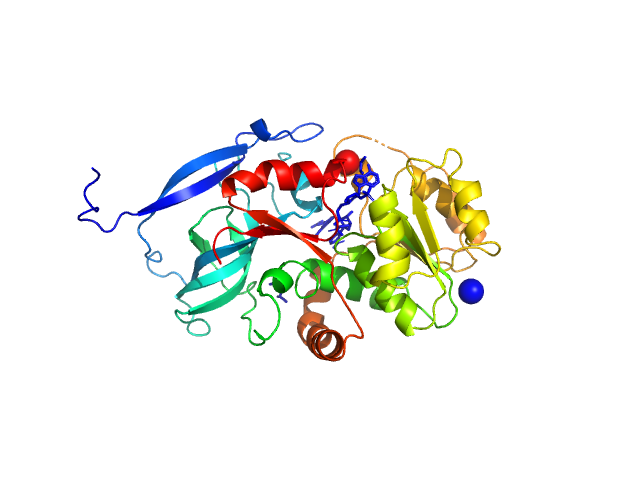
|
| Sample: |
Malus domestica double bond reductase dimer, 77 kDa Malus domestica protein
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl., pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2020 Oct 23
|
The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates
International Journal of Biological Macromolecules 171:89-99 (2021)
Caliandro R, Polsinelli I, Demitri N, Musiani F, Martens S, Benini S
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
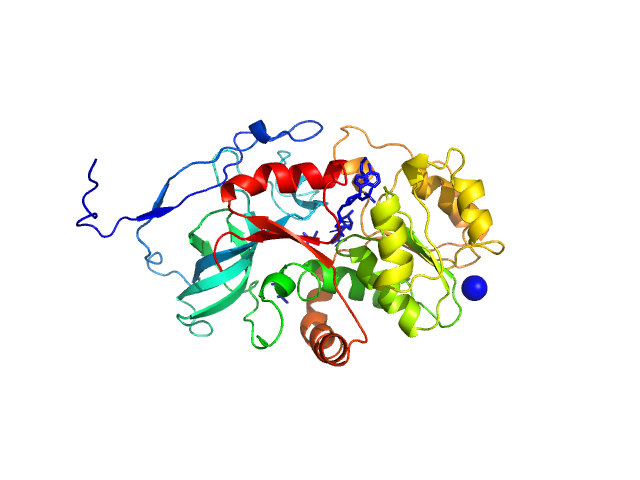
|
| Sample: |
Malus domestica double bond reductase dimer, 77 kDa Malus domestica protein
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5 mM NADPH, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2020 Oct 23
|
The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates
International Journal of Biological Macromolecules 171:89-99 (2021)
Caliandro R, Polsinelli I, Demitri N, Musiani F, Martens S, Benini S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|
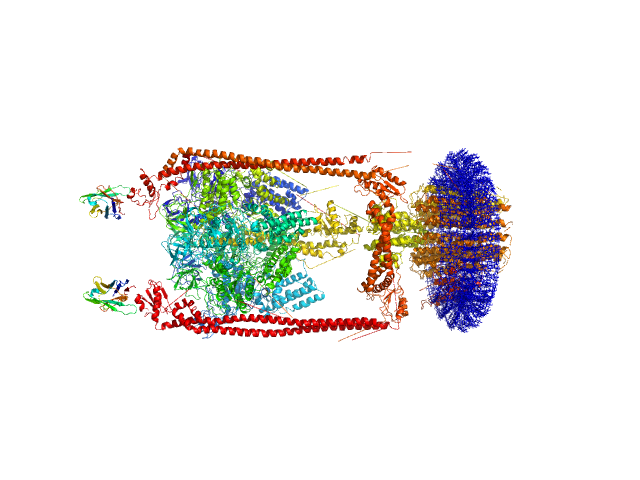
|
| Sample: |
N-Dodecyl-β-D-Maltopyranoside, 77 kDa synthetic construct
A-type ATP synthase monomer, 655 kDa Thermus thermophilus protein
Monoclonal Antibody Fragment dimer, 33 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris/HCl, 100 mM sucrose, 100 mM NaCl, 2 mM MgCl2, 10% glycerol, 0.05% b-DDM, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2013 Mar 3
|
MPBuilder: A PyMOL Plugin for Building and Refinement of Solubilized Membrane Proteins Against Small Angle X-ray Scattering Data
Journal of Molecular Biology :166888 (2021)
Molodenskiy D, Svergun D, Mertens H
|
| RgGuinier |
8.0 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
1549 |
nm3 |
|
|
|
|
|
|
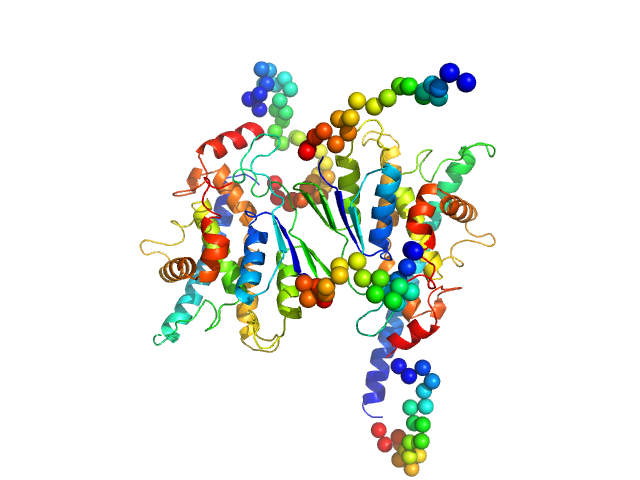
|
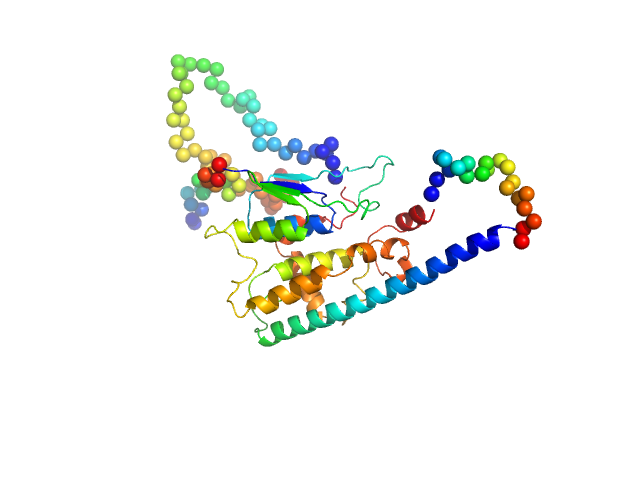
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆303-358 dimer, 70 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 28
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1-like 1 (∆125-143 isoform) monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM TRIS pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 10
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDKA4_fit1_model1.png)
|
| Sample: |
Properdin (dimer) dimer, 110 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 14
|
Properdin oligomers adopt rigid extended conformations supporting function.
Elife 10 (2021)
Pedersen DV, Pedersen MN, Mazarakis SM, Wang Y, Lindorff-Larsen K, Arleth L, Andersen GR
|
| RgGuinier |
8.1 |
nm |
| Dmax |
24.0 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDKB4_fit1_model1.png)
|
| Sample: |
Properdin (trimer) trimer, 165 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 14
|
Properdin oligomers adopt rigid extended conformations supporting function.
Elife 10 (2021)
Pedersen DV, Pedersen MN, Mazarakis SM, Wang Y, Lindorff-Larsen K, Arleth L, Andersen GR
|
| RgGuinier |
10.2 |
nm |
| Dmax |
27.0 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDKC4_fit1_model1.png)
|
| Sample: |
Properdin (tetramer) tetramer, 220 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 14
|
Properdin oligomers adopt rigid extended conformations supporting function.
Elife 10 (2021)
Pedersen DV, Pedersen MN, Mazarakis SM, Wang Y, Lindorff-Larsen K, Arleth L, Andersen GR
|
| RgGuinier |
13.1 |
nm |
| Dmax |
36.0 |
nm |
|
|
|
|
|
|
|
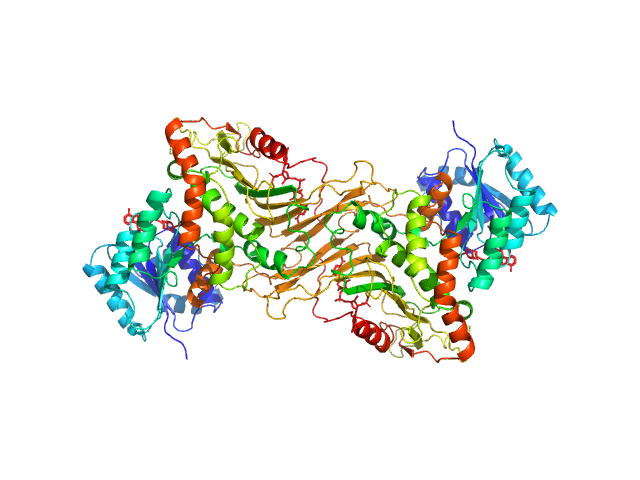
| Sample: |
Glucose-6-phosphate 1-dehydrogenase dimer, 119 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2019 Jul 24
|
Long-range structural defects by pathogenic mutations in most severe glucose-6-phosphate dehydrogenase deficiency
Proceedings of the National Academy of Sciences 118(4) (2021)
Horikoshi N, Hwang S, Gati C, Matsui T, Castillo-Orellana C, Raub A, Garcia A, Jabbarpour F, Batyuk A, Broweleit J, Xiang X, Chiang A, Broweleit R, Vöhringer-Martinez E, Mochly-Rosen D, Wakatsuki S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
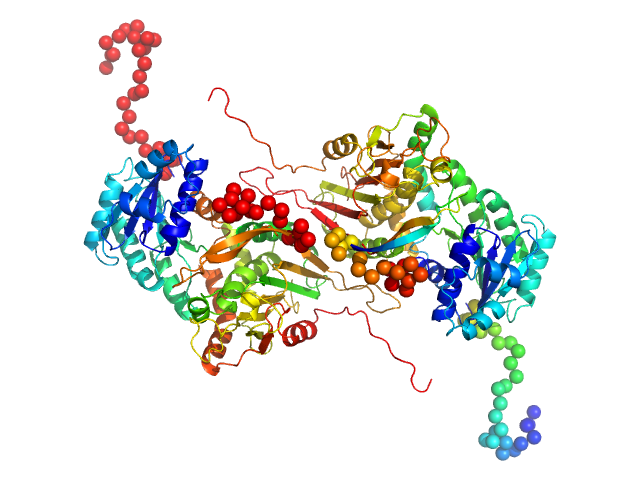
| Sample: |
Glucose-6-phosphate 1-dehydrogenase P396L dimer, 119 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2019 Jul 24
|
Long-range structural defects by pathogenic mutations in most severe glucose-6-phosphate dehydrogenase deficiency
Proceedings of the National Academy of Sciences 118(4) (2021)
Horikoshi N, Hwang S, Gati C, Matsui T, Castillo-Orellana C, Raub A, Garcia A, Jabbarpour F, Batyuk A, Broweleit J, Xiang X, Chiang A, Broweleit R, Vöhringer-Martinez E, Mochly-Rosen D, Wakatsuki S
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
178 |
nm3 |
|
|