|
|
|
|
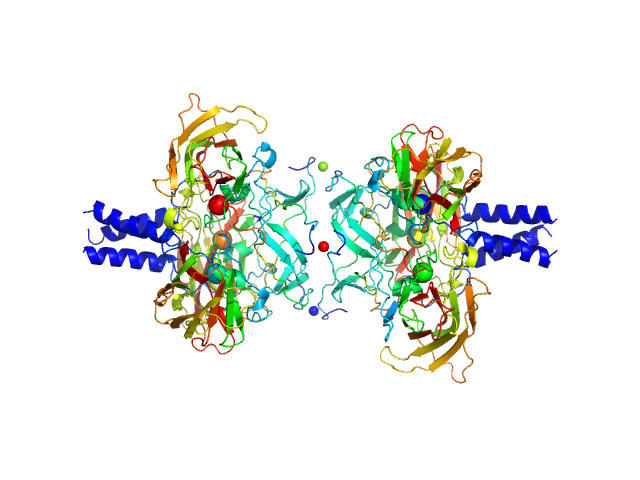
|
| Sample: |
SUN domain-containing protein 1 hexamer, 135 kDa Homo sapiens protein
Nesprin-4 hexamer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 21
|
A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife 10 (2021)
Gurusaran M, Davies OR
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
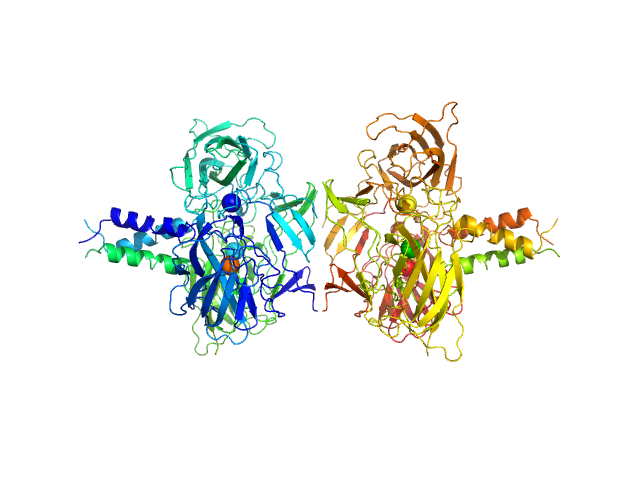
|
| Sample: |
SUN domain-containing protein 1 hexamer, 135 kDa Homo sapiens protein
Protein KASH5 hexamer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Dec 17
|
A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife 10 (2021)
Gurusaran M, Davies OR
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
244 |
nm3 |
|
|
|
|
|
|
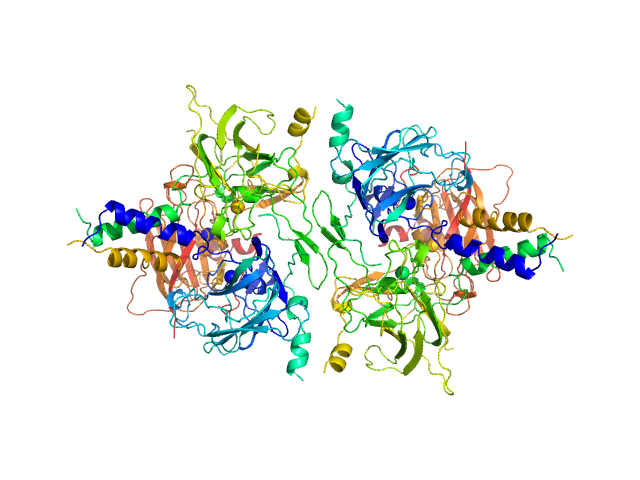
|
| Sample: |
SUN domain-containing protein 1 hexamer, 135 kDa Homo sapiens protein
Nesprin-1 hexamer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Dec 17
|
A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife 10 (2021)
Gurusaran M, Davies OR
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
254 |
nm3 |
|
|
|
|
|
|
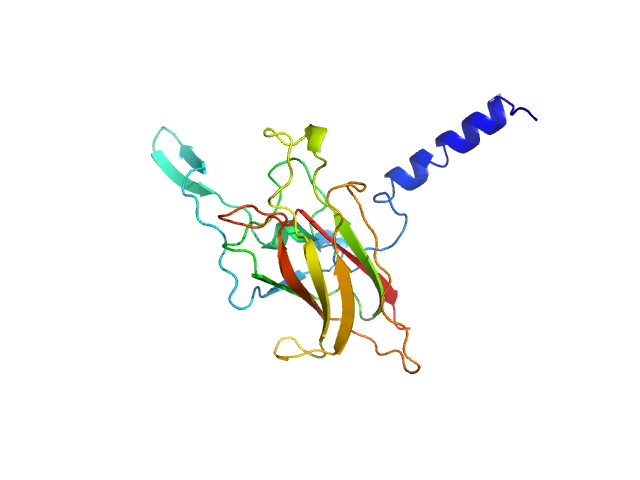
|
| Sample: |
SUN domain-containing protein 1, I673E monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jul 29
|
A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife 10 (2021)
Gurusaran M, Davies OR
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
24-mer double strand DNA from the GATA promoter dimer, 14 kDa synthetic oligonucleotide DNA
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 5
|
Biophysical characterization of the complex between the iron-responsive transcription factor Fep1 and DNA.
Eur Biophys J (2021)
Miele AE, Cervoni L, Le Roy A, Cutone A, Musci G, Ebel C, Bonaccorsi di Patti MC
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
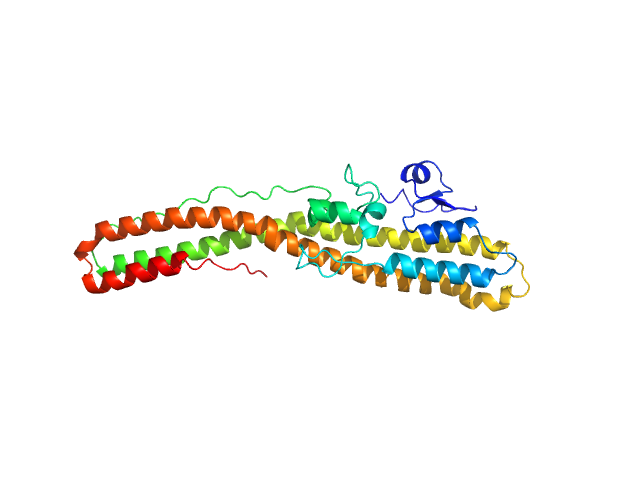
| Sample: |
Tetanus toxin monomer, 35 kDa Clostridium tetani protein
|
| Buffer: |
10 mM HEPES 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL45XU, SPring-8 on 2018 Oct 20
|
Structural flexibility of the tetanus neurotoxin revealed by crystallographic and solution scattering analyses.
J Struct Biol X 5:100045 (2021)
Zhang CM, Imoto Y, Hikima T, Inoue T
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
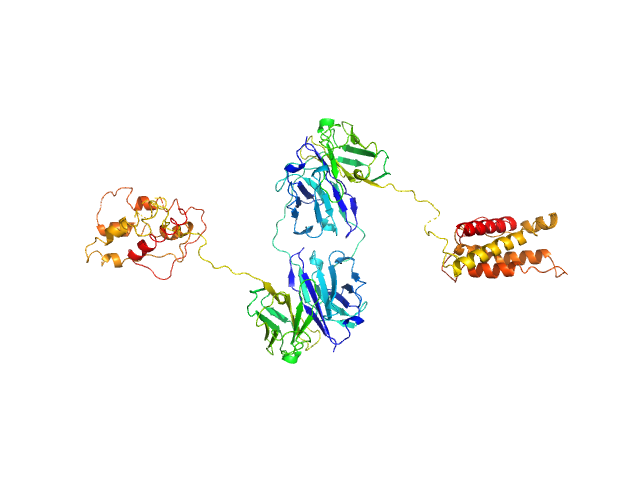
| Sample: |
L19-IL2 dimeric immunocytokine dimer, 84 kDa synthetic construct protein
|
| Buffer: |
25 mM HEPES/NaOH, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 23
|
Inference of molecular structure for characterization and improvement of clinical grade immunocytokines
Journal of Structural Biology :107696 (2021)
Ongaro T, Guarino S, Scietti L, Palamini M, Wulhfard S, Neri D, Villa A, Forneris F
|
| RgGuinier |
4.1 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|

|
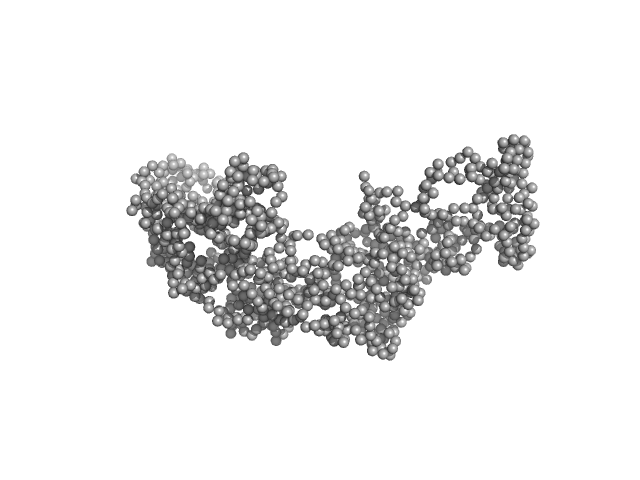
| Sample: |
L19L19-IL2 immunocytokine monomer, 69 kDa synthetic construct protein
|
| Buffer: |
25 mM HEPES/NaOH, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Mar 4
|
Inference of molecular structure for characterization and improvement of clinical grade immunocytokines
Journal of Structural Biology :107696 (2021)
Ongaro T, Guarino S, Scietti L, Palamini M, Wulhfard S, Neri D, Villa A, Forneris F
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
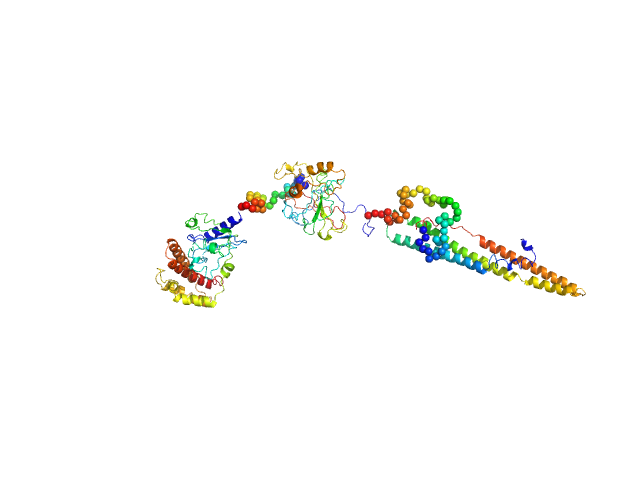
| Sample: |
IL12-L19L19 immunocytokine monomer, 112 kDa synthetic construct protein
|
| Buffer: |
25 mM HEPES/NaOH, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Mar 4
|
Inference of molecular structure for characterization and improvement of clinical grade immunocytokines
Journal of Structural Biology :107696 (2021)
Ongaro T, Guarino S, Scietti L, Palamini M, Wulhfard S, Neri D, Villa A, Forneris F
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
224 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ESX-1 secretion-associated protein EspK monomer, 27 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 20
|
Structural Analysis of the Partially Disordered Protein EspK from Mycobacterium Tuberculosis
Crystals 11(1):18 (2020)
Gijsbers A, Sánchez-Puig N, Gao Y, Peters P, Ravelli R, Siliqi D
|
| RgGuinier |
7.2 |
nm |
| Dmax |
3.3 |
nm |
| VolumePorod |
330 |
nm3 |
|
|