|
|
|
|
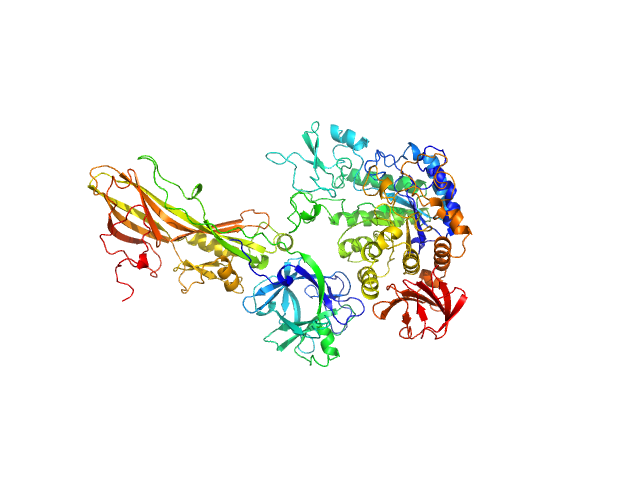
|
| Sample: |
Binary larvicide subunit BinB monomer, 53 kDa Lysinibacillus sphaericus protein
Synthetic construct (mutant, His-tagged): Mosquito-larvicidal BinAB toxin receptor protein (Neutral and basic amino acid transport protein rBAT) monomer, 65 kDa Culex quinquefasciatus protein
|
| Buffer: |
PBS buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl), pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2020 Feb 24
|
Liposome-Based Study Provides Insight into Cellular Internalization Mechanism of Mosquito-Larvicidal BinAB Toxin.
J Membr Biol (2020)
Sharma M, Kumar A, Kumar V
|
| RgGuinier |
4.6 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Binary larvicide subunit BinB monomer, 53 kDa Lysinibacillus sphaericus protein
Synthetic construct (mutant, His-tagged): Mosquito-larvicidal BinAB toxin receptor protein (Neutral and basic amino acid transport protein rBAT) monomer, 65 kDa Culex quinquefasciatus protein
|
| Buffer: |
25 mM HEPES, pH 7.5, 25 mM NaCl, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-I facility, Dhruva Reactor, Bhabha Atomic Research Centre on 2019 Apr 26
|
Liposome-Based Study Provides Insight into Cellular Internalization Mechanism of Mosquito-Larvicidal BinAB Toxin.
J Membr Biol (2020)
Sharma M, Kumar A, Kumar V
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
174 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Major tail protein monomer, 40 kDa Salmonella virus Chi protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.03 % NaN3, 5.0 % glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Mar 8
|
The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun 11(1):3748 (2020)
Hardy JM, Dunstan RA, Grinter R, Belousoff MJ, Wang J, Pickard D, Venugopal H, Dougan G, Lithgow T, Coulibaly F
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
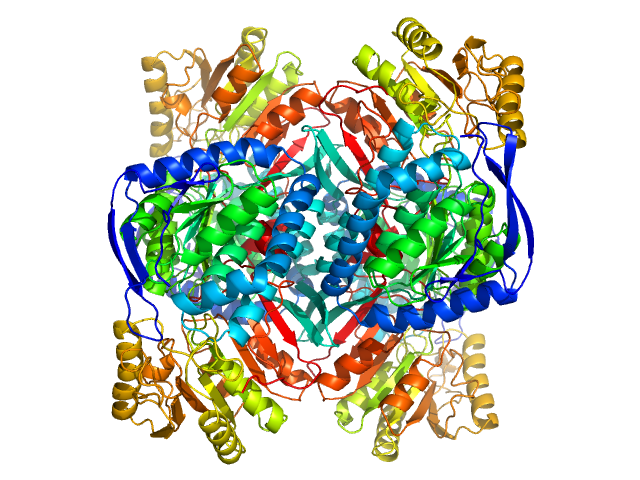
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta-ketoacyl synthase Bamb_5924 dimer, 10 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 6
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|

|
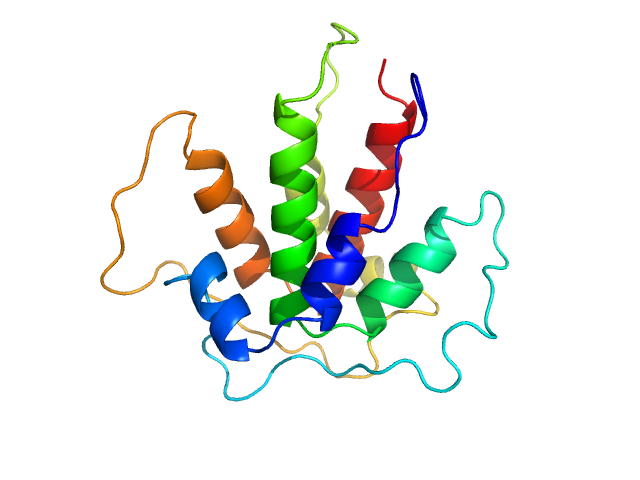
| Sample: |
Beta-ketoacyl synthase Bamb_5920 CDD/Bamb_5919 NDD artificial protein fusion dimer, 13 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-ketoacyl synthase Bamb_5925 CDD/Bamb_5924 NDD artificial protein fusion dimer, 18 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
77 |
nm3 |
|
|