|
|
|
|
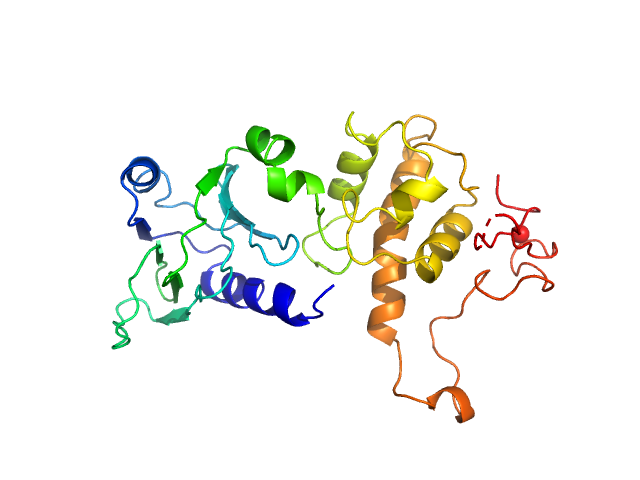
|
| Sample: |
Endonuclease 8 monomer, 31 kDa Escherichia coli protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 2% glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Mar 9
|
Unique Structural Features of Mammalian NEIL2 DNA Glycosylase Prime Its Activity for Diverse DNA Substrates and Environments.
Structure (2020)
Eckenroth BE, Cao VB, Averill AM, Dragon JA, Doublié S
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
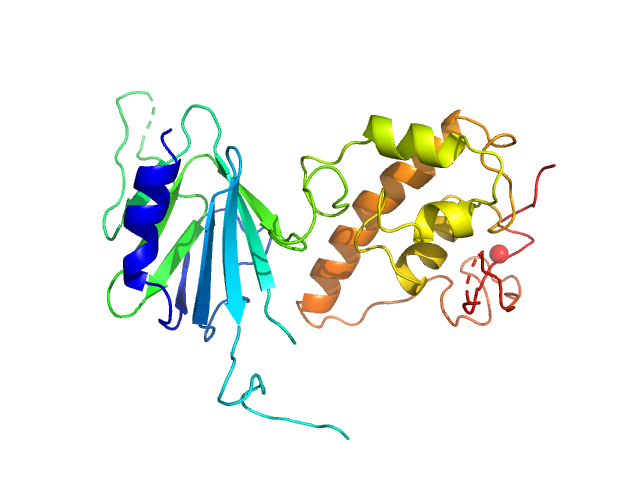
|
| Sample: |
Nei like DNA glycosylase 2 (Δ67-133) monomer, 33 kDa Monodelphis domestica protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 2% glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Mar 9
|
Unique Structural Features of Mammalian NEIL2 DNA Glycosylase Prime Its Activity for Diverse DNA Substrates and Environments.
Structure (2020)
Eckenroth BE, Cao VB, Averill AM, Dragon JA, Doublié S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
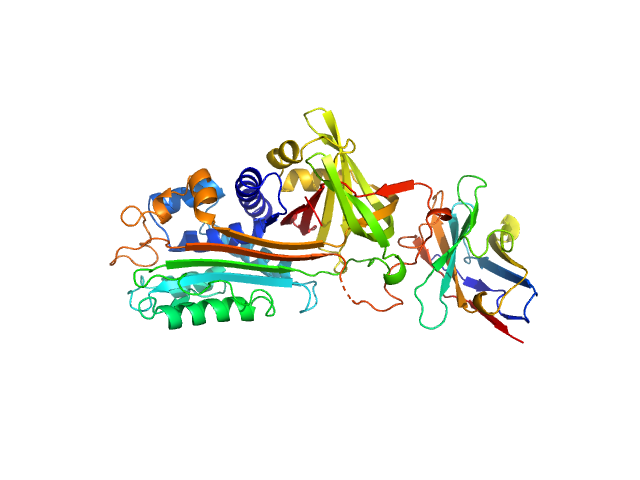
|
| Sample: |
Plasminogen activator inhibitor 1 monomer, 43 kDa Homo sapiens protein
VHH-s-a93 (Ig module Nb93) monomer, 13 kDa Vicugna pacos protein
|
| Buffer: |
30 mM BIS-TRIS pH 5.5, 300 mM sodium chloride, 5% v/v glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 5
|
Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity
International Journal of Molecular Sciences 21(16):5859 (2020)
Sillen M, Weeks S, Strelkov S, Declerck P
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
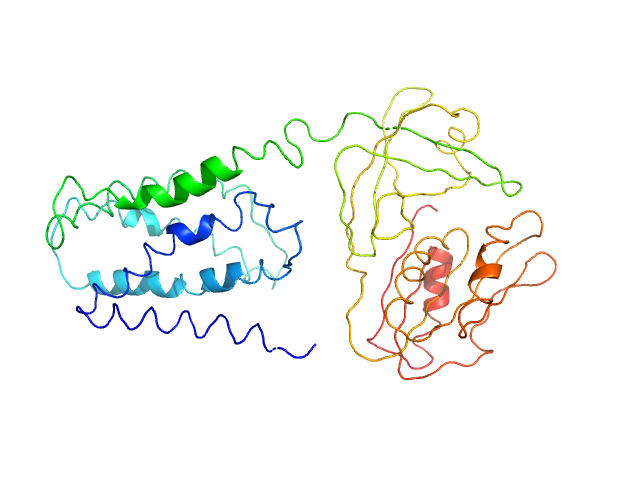
|
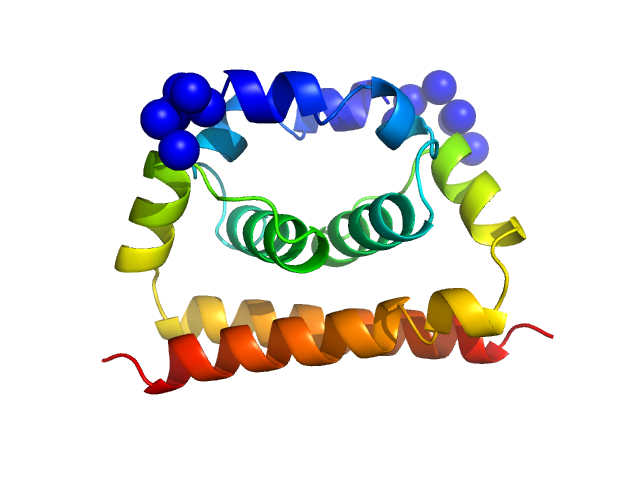
| Sample: |
FAD-binding FR-type domain-containing protein monomer, 47 kDa Streptococcus pneumoniae protein
|
| Buffer: |
50 mM Tris-HCl pH 7, 300 mM NaCl, 5 mM LMNG, 10 µM FAD, 21.4% D₂O, pH: 7 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Jun 26
|
Interdomain Flexibility within NADPH Oxidase Suggested by SANS Using LMNG Stealth Carrier.
Biophys J 119(3):605-618 (2020)
Vermot A, Petit-Härtlein I, Breyton C, Le Roy A, Thépaut M, Vivès C, Moulin M, Härtlein M, Grudinin S, Smith SME, Ebel C, Martel A, Fieschi F
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
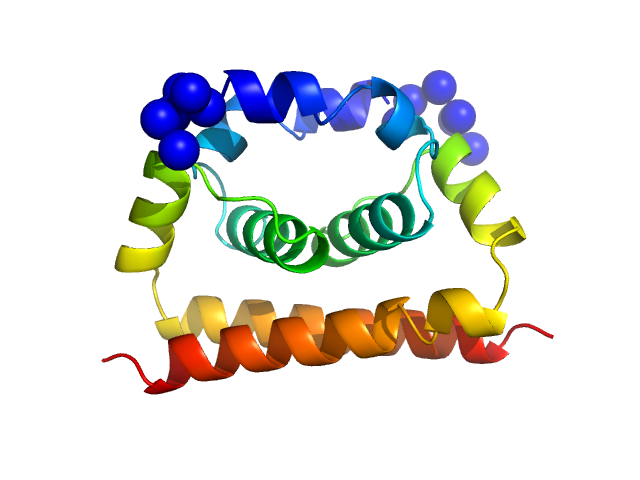
| Sample: |
Dengue Virus 2 New Guinea C dimer, 19 kDa Dengue virus 2 protein
|
| Buffer: |
100 mM NaCl, 25 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2018 Jul 5
|
A cocrystal structure of dengue capsid protein in complex of inhibitor.
Proc Natl Acad Sci U S A 117(30):17992-18001 (2020)
Xia H, Xie X, Zou J, Noble CG, Russell WK, Holthauzen LMF, Choi KH, White MA, Shi PY
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
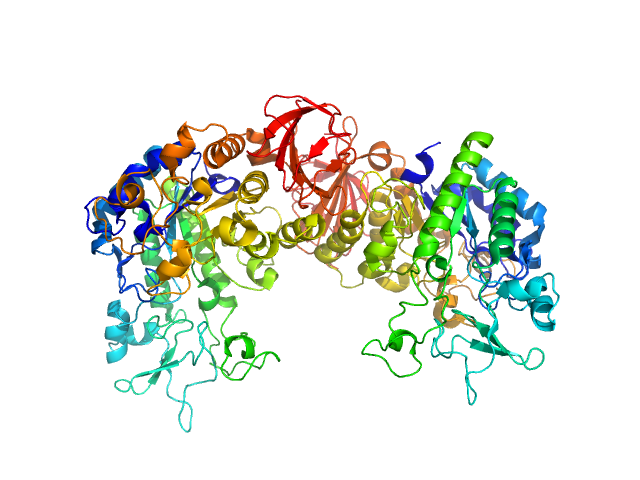
| Sample: |
Dengue Virus 2 New Guinea C dimer, 19 kDa Dengue virus 2 protein
|
| Buffer: |
100 mM NaCl, 25 mM HEPES, pH 7.4, 10 µM ST148, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2018 Jul 26
|
A cocrystal structure of dengue capsid protein in complex of inhibitor.
Proc Natl Acad Sci U S A 117(30):17992-18001 (2020)
Xia H, Xie X, Zou J, Noble CG, Russell WK, Holthauzen LMF, Choi KH, White MA, Shi PY
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
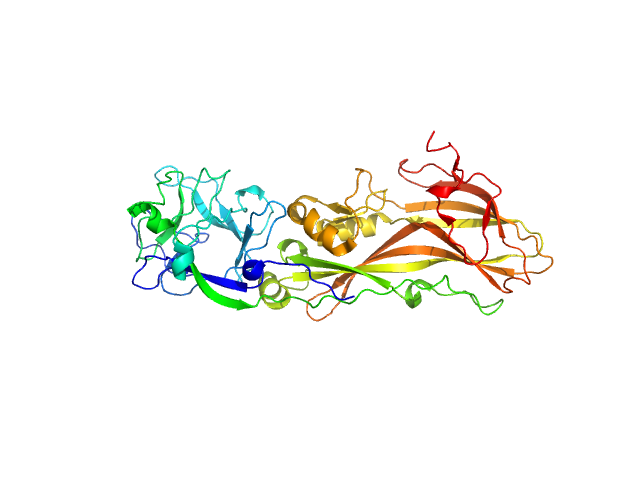
| Sample: |
Synthetic construct (mutant, His-tagged): Mosquito-larvicidal BinAB toxin receptor protein (Neutral and basic amino acid transport protein rBAT) dimer, 129 kDa Culex quinquefasciatus protein
|
| Buffer: |
PBS buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl), pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2020 Feb 24
|
Liposome-Based Study Provides Insight into Cellular Internalization Mechanism of Mosquito-Larvicidal BinAB Toxin.
J Membr Biol (2020)
Sharma M, Kumar A, Kumar V
|
| RgGuinier |
4.0 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Binary larvicide subunit BinB monomer, 53 kDa Lysinibacillus sphaericus protein
|
| Buffer: |
PBS buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl), pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2020 Feb 24
|
Liposome-Based Study Provides Insight into Cellular Internalization Mechanism of Mosquito-Larvicidal BinAB Toxin.
J Membr Biol (2020)
Sharma M, Kumar A, Kumar V
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
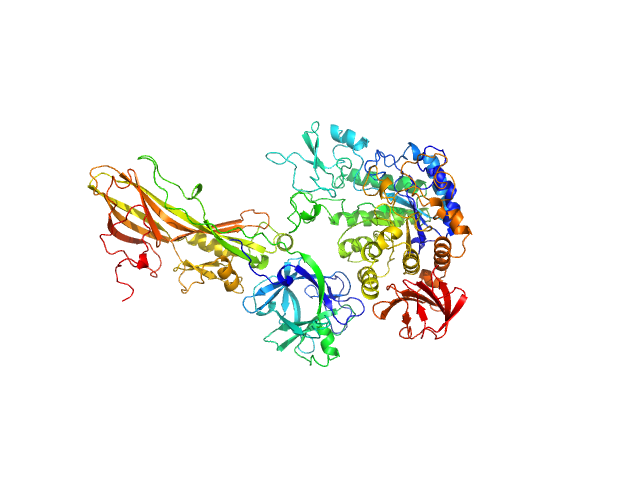
| Sample: |
Binary larvicide subunit BinB monomer, 53 kDa Lysinibacillus sphaericus protein
Synthetic construct (mutant, His-tagged): Mosquito-larvicidal BinAB toxin receptor protein (Neutral and basic amino acid transport protein rBAT) monomer, 65 kDa Culex quinquefasciatus protein
|
| Buffer: |
PBS buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl), pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2020 Feb 24
|
Liposome-Based Study Provides Insight into Cellular Internalization Mechanism of Mosquito-Larvicidal BinAB Toxin.
J Membr Biol (2020)
Sharma M, Kumar A, Kumar V
|
| RgGuinier |
4.6 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
|
|
|
|
|
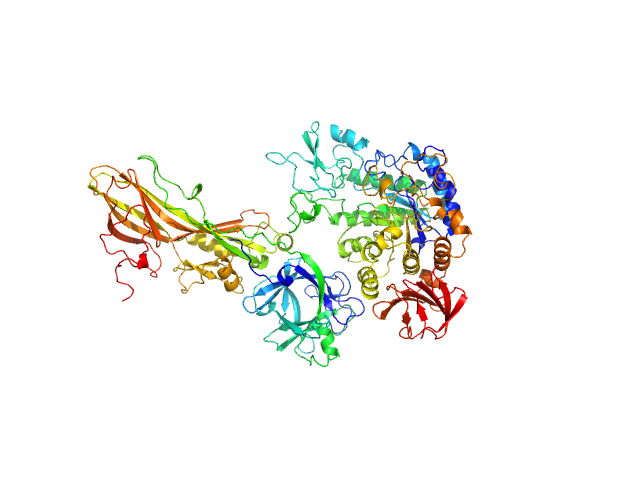
| Sample: |
Binary larvicide subunit BinB monomer, 53 kDa Lysinibacillus sphaericus protein
Synthetic construct (mutant, His-tagged): Mosquito-larvicidal BinAB toxin receptor protein (Neutral and basic amino acid transport protein rBAT) monomer, 65 kDa Culex quinquefasciatus protein
|
| Buffer: |
25 mM HEPES, pH 7.5, 25 mM NaCl, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-I facility, Dhruva Reactor, Bhabha Atomic Research Centre on 2019 Apr 26
|
Liposome-Based Study Provides Insight into Cellular Internalization Mechanism of Mosquito-Larvicidal BinAB Toxin.
J Membr Biol (2020)
Sharma M, Kumar A, Kumar V
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
174 |
nm3 |
|
|