|
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
DNA (TTTTTACTCC) monomer, 1 kDa DNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 May 27
|
TIA-1 RRM23 binding and recognition of target oligonucleotides.
Nucleic Acids Res 45(8):4944-4957 (2017)
Waris S, García-Mauriño SM, Sivakumaran A, Beckham SA, Loughlin FE, Gorospe M, Díaz-Moreno I, Wilce MCJ, Wilce JA
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
RNA (UUUUUACU) monomer, 1 kDa RNA
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 May 27
|
TIA-1 RRM23 binding and recognition of target oligonucleotides.
Nucleic Acids Res 45(8):4944-4957 (2017)
Waris S, García-Mauriño SM, Sivakumaran A, Beckham SA, Loughlin FE, Gorospe M, Díaz-Moreno I, Wilce MCJ, Wilce JA
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
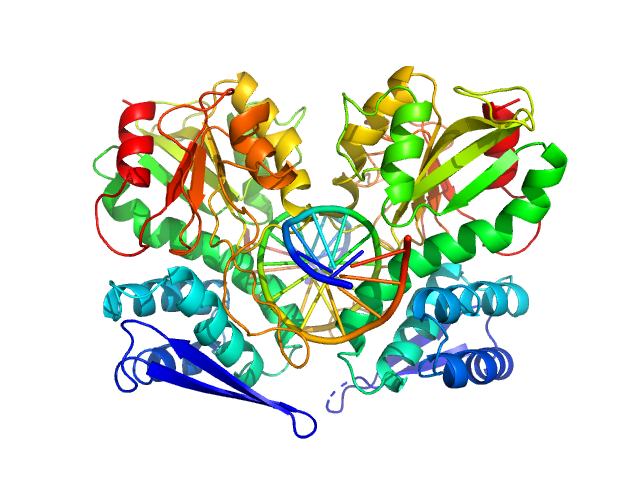
| Sample: |
Type-2 restriction enzyme AgeI dimer, 61 kDa Thalassobius gelatinovorus protein
Cognate DNA oligoduplex with 5'-T overhang dimer, 8 kDa DNA
|
| Buffer: |
10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM CaCl₂, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 14
|
Restriction endonuclease AgeI is a monomer which dimerizes to cleave DNA.
Nucleic Acids Res 45(6):3547-3558 (2017)
Tamulaitiene G, Jovaisaite V, Tamulaitis G, Songailiene I, Manakova E, Zaremba M, Grazulis S, Xu SY, Siksnys V
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
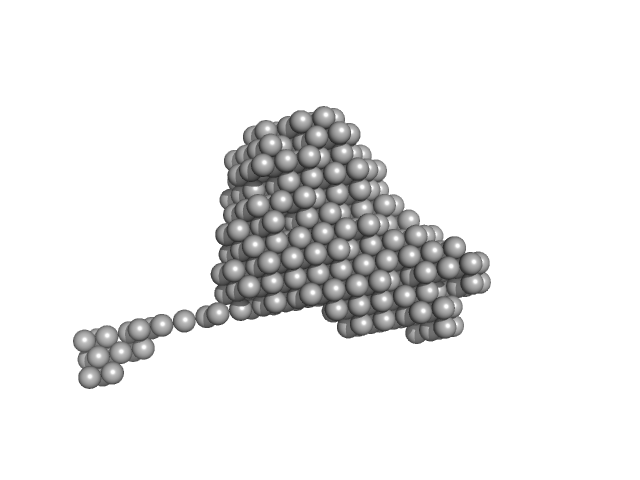
| Sample: |
Endonuclease 8-like 1 monomer, 45 kDa Homo sapiens protein
DsDNA monomer, 2 kDa DNA
Proliferating cell nuclear antigen monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
25mM HEPES 100mM NaCl 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Jan 20
|
Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase.
Nucleic Acids Res 45(5):2897-2909 (2017)
Prakash A, Moharana K, Wallace SS, Doublié S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
|
|
|
|
|
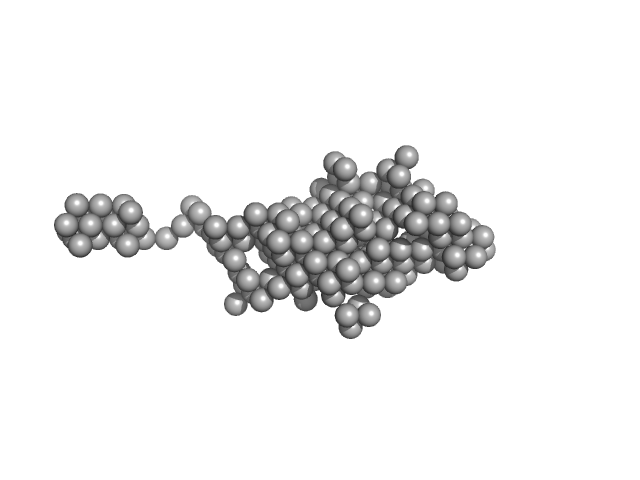
| Sample: |
Endonuclease 8-like 1 monomer, 45 kDa Homo sapiens protein
DsDNA monomer, 2 kDa DNA
|
| Buffer: |
25mM HEPES 100mM NaCl 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Jan 20
|
Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase.
Nucleic Acids Res 45(5):2897-2909 (2017)
Prakash A, Moharana K, Wallace SS, Doublié S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleasome Core Particle with Widom 601 DNA - HISTONE H2A-H2B Heterodimer dimer, 48 kDa Xenopus laevis protein
Nucleasome Core Particle with Widom 601 DNA - HISTONE H3-H4 Heterodimer dimer, 46 kDa Xenopus laevis protein
Nucleasome Core Particle with Widom 601 DNA - dsDNA monomer, 92 kDa Xenopus laevis DNA
|
| Buffer: |
20 mM Tris-Cl, 0.1 mM EDTA, 0.1 mM DTT, 50% sucrose, 1.2 M NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Apr 14
|
Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core.
Proc Natl Acad Sci U S A 114(2):334-339 (2017)
Chen Y, Tokuda JM, Topping T, Meisburger SP, Pabit SA, Gloss LM, Pollack L
|
|
|
|
|
|
|
|
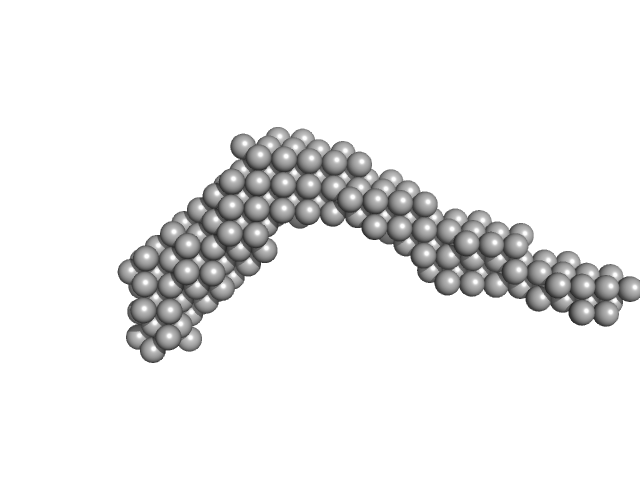
| Sample: |
Nucleasome Core Particle with Widom 601 DNA - HISTONE H2A-H2B Heterodimer dimer, 48 kDa Xenopus laevis protein
Nucleasome Core Particle with Widom 601 DNA - HISTONE H3-H4 Heterodimer dimer, 46 kDa Xenopus laevis protein
Nucleasome Core Particle with Widom 601 DNA - dsDNA monomer, 92 kDa Xenopus laevis DNA
|
| Buffer: |
20 mM Tris-Cl, 0.1 mM EDTA, 0.1 mM DTT, 50% sucrose, 1.9 M NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Apr 14
|
Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core.
Proc Natl Acad Sci U S A 114(2):334-339 (2017)
Chen Y, Tokuda JM, Topping T, Meisburger SP, Pabit SA, Gloss LM, Pollack L
|
|
|
|
|
|
|
|
| Sample: |
AIR-3A tetramer, 26 kDa RNA
Interleukin-6 receptor subunit alpha dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2014 Oct 1
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
6.7 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
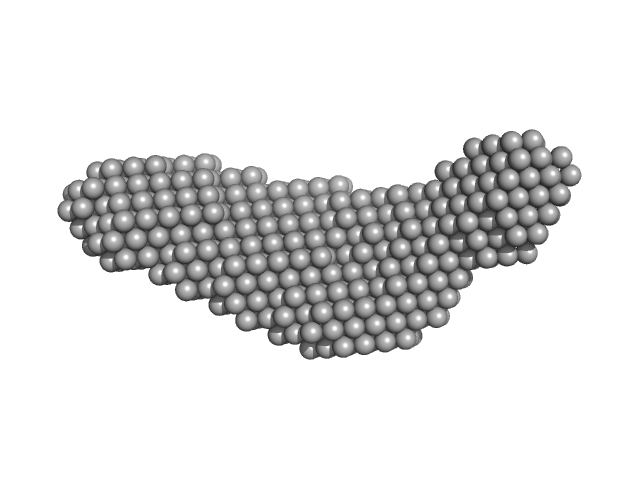
|
| Sample: |
Pre-B-cell leukemia transcription factor 1 monomer, 34 kDa Homo sapiens protein
Homeobox protein PKNOX1 monomer, 38 kDa Homo sapiens protein
DNA 22mer monomer, 12 kDa DNA
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 5% glycerol 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
The flexibility of a homeodomain transcription factor heterodimer and its allosteric regulation by DNA binding.
FEBS J 283(16):3134-54 (2016)
Mathiasen L, Valentini E, Boivin S, Cattaneo A, Blasi F, Svergun DI, Bruckmann C
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
173 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureobox dsDNA monomer, 13 kDa synthetic construct DNA
Aureochrome 1a bZIP-LOV module dimer, 57 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
50 mM Tris 50 mM boric acid 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 6
|
Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes.
Nucleic Acids Res 44(12):5957-70 (2016)
Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kroth PG, Essen LO, Kottke T
|
| RgGuinier |
4.5 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
97 |
nm3 |
|
|