|
|
|
|
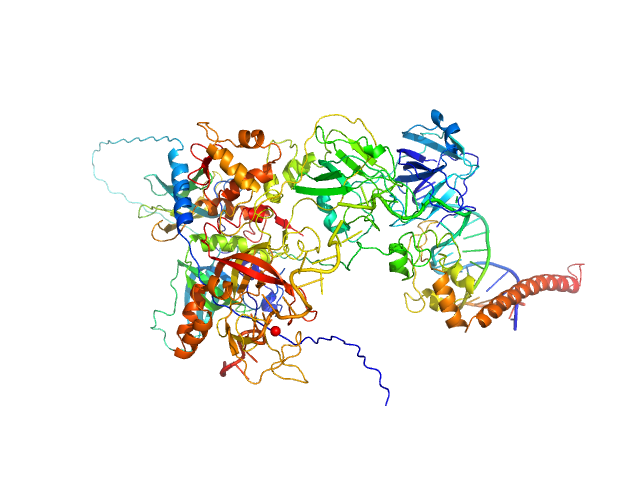
|
| Sample: |
DNA repair protein complementing XP-A cells monomer, 27 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 49 kDa Homo sapiens protein
Replication protein A 32 kDa subunit monomer, 25 kDa Homo sapiens protein
Replication protein A 14 kDa subunit monomer, 14 kDa Homo sapiens protein
3-prime ss-ds DNA junction NER model substrate monomer, 17 kDa DNA
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Mar 4
|
Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair
Proceedings of the National Academy of Sciences 119(34) (2022)
Kim M, Kim H, D’Souza A, Gallagher K, Jeong E, Topolska-Wós A, Ogorodnik Le Meur K, Tsai C, Tsai M, Kee M, Tainer J, Yeo J, Chazin W, Schärer O
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|
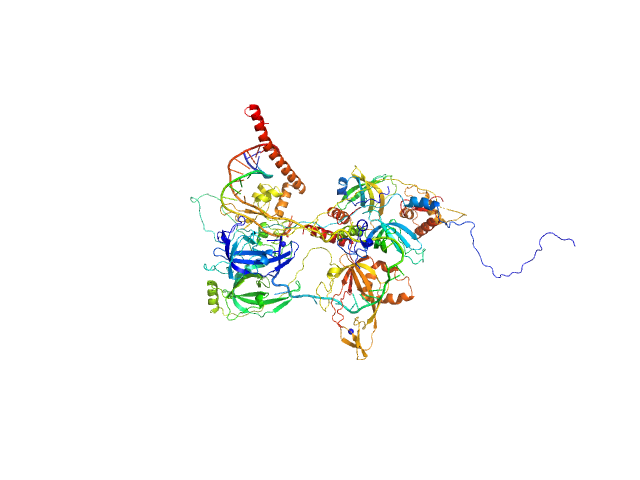
|
| Sample: |
Replication protein A 14 kDa subunit monomer, 14 kDa Homo sapiens protein
DNA repair protein complementing XP-A cells monomer, 27 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 49 kDa Homo sapiens protein
Replication protein A 32 kDa subunit monomer, 25 kDa Homo sapiens protein
5-prime ss-ds DNA junction NER model substrate monomer, 17 kDa DNA
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Mar 4
|
Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair
Proceedings of the National Academy of Sciences 119(34) (2022)
Kim M, Kim H, D’Souza A, Gallagher K, Jeong E, Topolska-Wós A, Ogorodnik Le Meur K, Tsai C, Tsai M, Kee M, Tainer J, Yeo J, Chazin W, Schärer O
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Transcription factor p65 19-321 monomer, 35 kDa Mus musculus protein
IFN kB DNA monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
237 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Transcription factor p65 19-321 monomer, 35 kDa Mus musculus protein
Urokinase kB monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
244 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transcription factor p65 19-549 monomer, 58 kDa Mus musculus protein
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
IFN kB DNA monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 4
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
314 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transcription factor p65 19-549 monomer, 58 kDa Mus musculus protein
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Urokinase kB monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 4
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
201 |
nm3 |
|
|
|
|
|
|
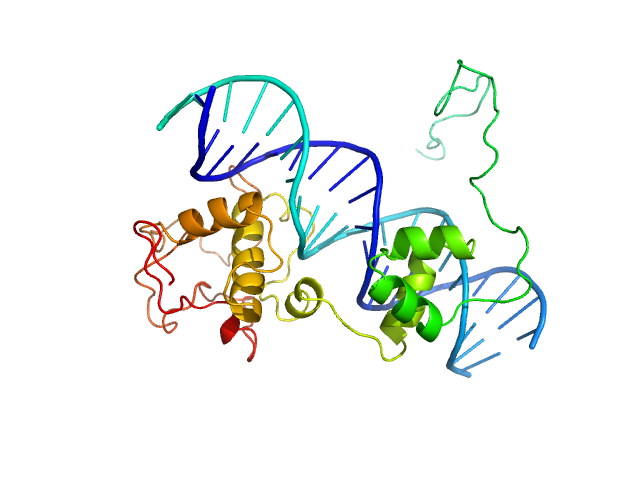
|
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
21mer dsDNA monomer, 13 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
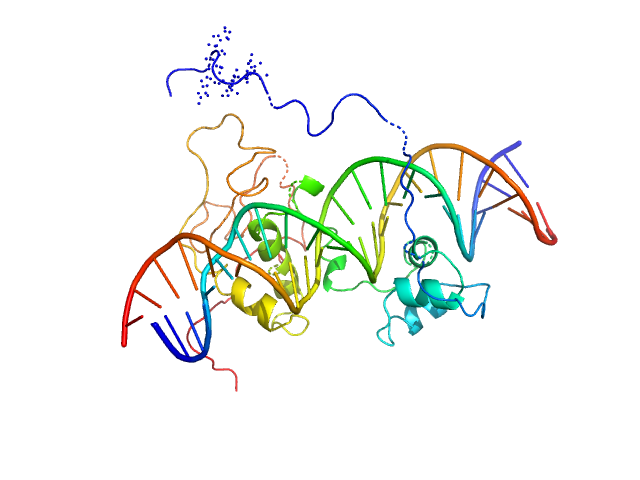
|
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
24mer dsDNA monomer, 15 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
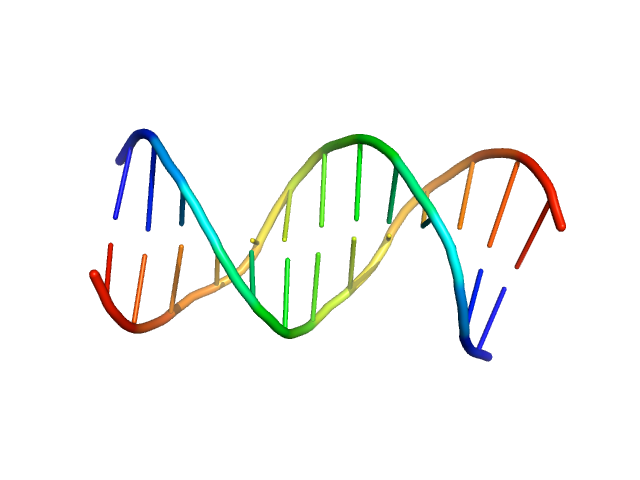
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
13mer dsDNA monomer, 8 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
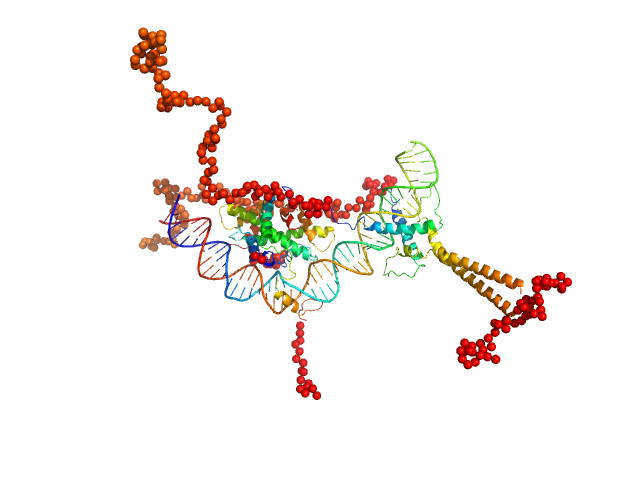
| Sample: |
Upstream stimulatory factor 1 dimer, 50 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 48bp monomer, 30 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
155 |
nm3 |
|
|