|
|
|
|

|
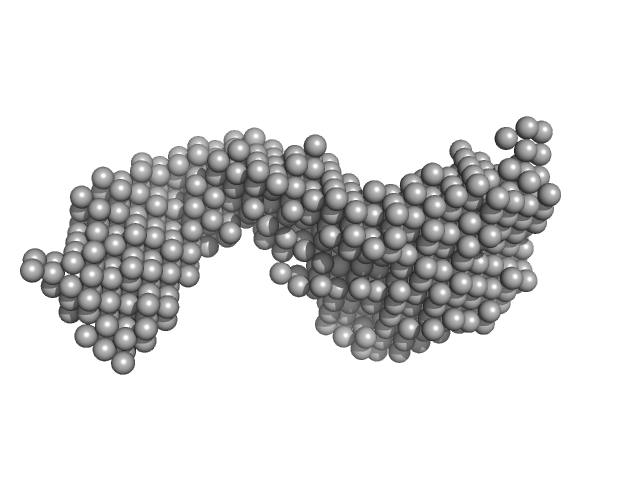
| Sample: |
Upstream stimulatory factor 1 dimer, 50 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 50bp monomer, 31 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein max (Isoform 2, short, 13-21: missing) dimer, 39 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 48bp monomer, 30 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Dec 10
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Brain tumor protein monomer, 32 kDa Drosophila melanogaster protein
Maternal protein pumilio monomer, 38 kDa Drosophila melanogaster protein
Protein nanos monomer, 11 kDa Drosophila melanogaster protein
Hunchback mRNA Nanos Response Element 2 monomer, 7 kDa Drosophila melanogaster RNA
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM DTT, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 12
|
Structure and dynamics of the quaternary hunchback mRNA translation repression complex.
Nucleic Acids Res 49(15):8866-8885 (2021)
Macošek J, Simon B, Linse JB, Jagtap PKA, Winter SL, Foot J, Lapouge K, Perez K, Rettel M, Ivanović MT, Masiewicz P, Murciano B, Savitski MM, Loedige I, Hub JS, Gabel F, Hennig J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoid-related orphan receptor-gamma monomer, 56 kDa Homo sapiens protein
Classic-RORgamma Response Element dimer, 19 kDa Homo sapiens DNA
|
| Buffer: |
25 mM HEPES, 150 mM TCEP, 2% Glycerol, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Nov 20
|
Conformational Changes of RORγ During Response Element Recognition and Coregulator Engagement
Journal of Molecular Biology :167258 (2021)
Strutzenberg T, Zhu Y, Novick S, Garcia-Ordonez R, Doebelin C, He Y, Ra Chang M, Kamenecka T, Edwards D, Griffin P
|
| RgGuinier |
5.5 |
nm |
| Dmax |
22.9 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoid-related orphan receptor-gamma monomer, 56 kDa Homo sapiens protein
Variant-RORgamma Response Element dimer, 18 kDa Homo sapiens DNA
|
| Buffer: |
25 mM HEPES, 150 mM TCEP, 2% Glycerol, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Nov 20
|
Conformational Changes of RORγ During Response Element Recognition and Coregulator Engagement
Journal of Molecular Biology :167258 (2021)
Strutzenberg T, Zhu Y, Novick S, Garcia-Ordonez R, Doebelin C, He Y, Ra Chang M, Kamenecka T, Edwards D, Griffin P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
22.1 |
nm |
| VolumePorod |
112 |
nm3 |
|
|
|
|
|
|
|
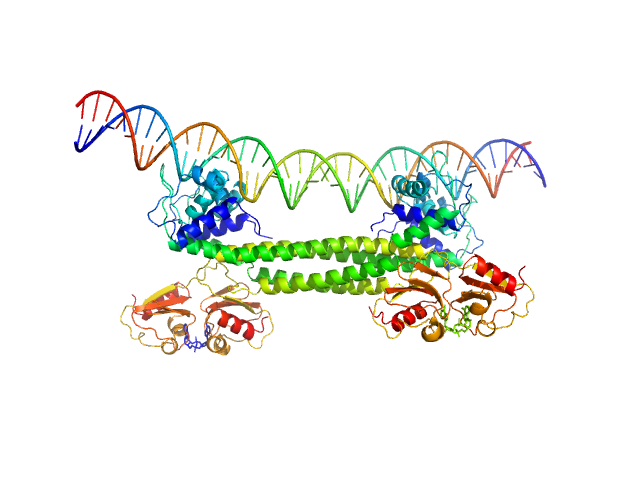
| Sample: |
Transcriptional repressor BusR tetramer, 95 kDa Streptococcus agalactiae protein
BusR Recognition sequence monomer, 28 kDa synthetic construct DNA
|
| Buffer: |
20mM HEPES, pH6.5, 100mM NaCl, 3% glycerol (v/v), pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res (2021)
Bandera AM, Bartho J, Lammens K, Drexler DJ, Kleinschwärzer J, Hopfner KP, Witte G
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
|
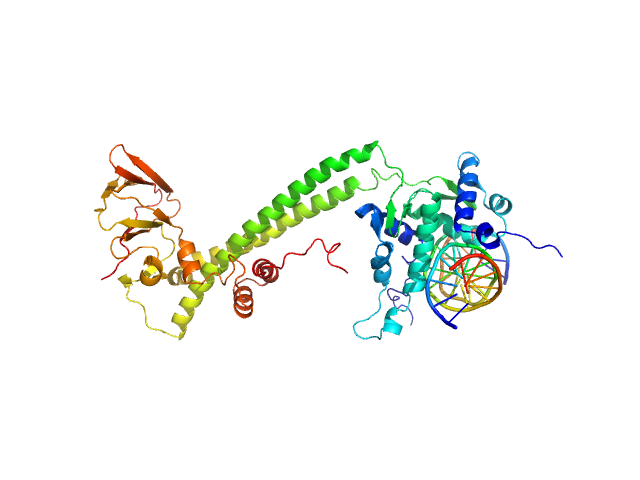
| Sample: |
Transcription factor E2F1 monomer, 20 kDa Homo sapiens protein
15-mer DNA oligo of the human E2F1 promoter monomer, 9 kDa DNA
E2F dimerization partner 1 monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-Cl pH 8.0, 150 mM NaCl and 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 19
|
High Conformational Flexibility of the E2F1/DP1/DNA Complex.
J Mol Biol 433(18):167119 (2021)
Saad D, Paissoni C, Chaves-Sanjuan A, Nardini M, Mantovani R, Gnesutta N, Camilloni C
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Signal recognition particle 9 monomer, 12 kDa Plasmodium falciparum protein
Signal recognition particle 14 monomer, 12 kDa Plasmodium falciparum protein
Full-length SRP Alu RNA monomer, 38 kDa Plasmodium falciparum RNA
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM KCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 22
|
Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation.
Commun Biol 4(1):600 (2021)
Soni K, Kempf G, Manalastas-Cantos K, Hendricks A, Flemming D, Guizetti J, Simon B, Frischknecht F, Svergun DI, Wild K, Sinning I
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Signal recognition particle 9 monomer, 12 kDa Plasmodium falciparum protein
Signal recognition particle 14 monomer, 12 kDa Plasmodium falciparum protein
SRP Alu RNA 5' domain monomer, 24 kDa Plasmodium falciparum RNA
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM KCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 22
|
Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation.
Commun Biol 4(1):600 (2021)
Soni K, Kempf G, Manalastas-Cantos K, Hendricks A, Flemming D, Guizetti J, Simon B, Frischknecht F, Svergun DI, Wild K, Sinning I
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histone H3 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B monomer, 14 kDa Xenopus laevis protein
Non-linker Ended Trinucleosome DNA monomer, 172 kDa DNA
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 1 mM DTT 50% w/v sucrose, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Mar 11
|
Solution structure(s) of trinucleosomes from contrast variation SAXS
Nucleic Acids Research (2021)
Mauney A, Muthurajan U, Luger K, Pollack L
|
| RgGuinier |
12.9 |
nm |
| Dmax |
41.8 |
nm |
|
|