|
|
|
|
|
| Sample: |
Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 monomer, 18 kDa Saccharomyces cerevisiae (strain … protein
Short RNA oligonucleotide (NPL3 binding sequence) monomer, 4 kDa RNA
|
| Buffer: |
20 mM NaPO4, 50 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2020 Feb 20
|
Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res (2022)
Keil P, Wulf A, Kachariya N, Reuscher S, Hühn K, Silbern I, Altmüller J, Keller M, Stehle R, Zarnack K, Sattler M, Urlaub H, Sträßer K
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
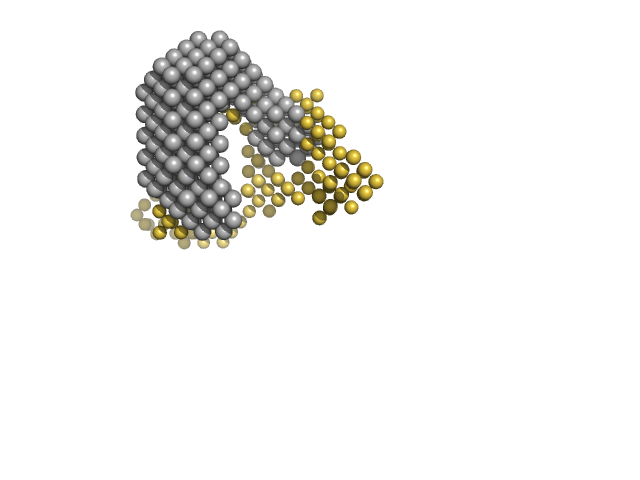
|
| Sample: |
Multidrug resistance operon repressor dimer, 32 kDa Pseudomonas aeruginosa protein
34 base pair double-stranded DNA monomer, 21 kDa synthetic construct DNA
|
| Buffer: |
20mM NaPO4, 150 mM NaCl, 10 mM DTT, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 May 30
|
Small-angle X-ray and neutron scattering of MexR and its complex with DNA supports a conformational selection binding model
Biophysical Journal (2022)
Caporaletti F, Pietras Z, Morad V, Mårtensson L, Gabel F, Wallner B, Martel A, Sunnerhagen M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|

|
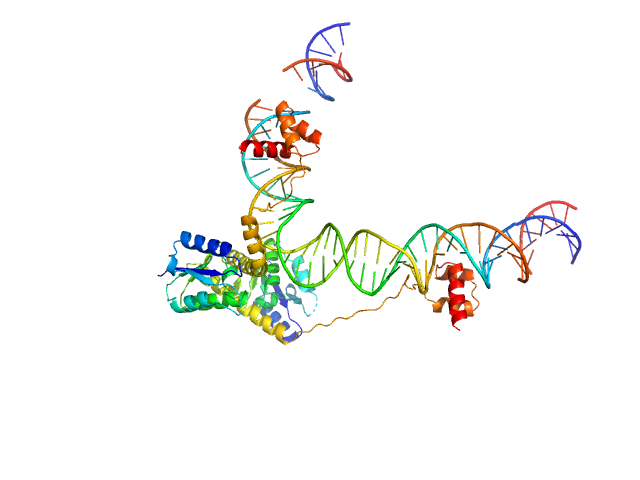
| Sample: |
Probable exodeoxyribonuclease III protein XthA monomer, 31 kDa Mycobacterium tuberculosis protein
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
Beta sliding clamp dimer, 86 kDa Mycobacterium tuberculosis protein
DNA ligase A nicked DNA substrate dimer, 16 kDa DNA
|
| Buffer: |
50 mM Tris pH 8.0, 200 mM NaCl , 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 17
|
Regulation of futile ligation during early steps of BER in M. tuberculosis is carried out by a β-clamp-XthA-LigA tri-component complex
International Journal of Biological Macromolecules (2022)
Shukla A, Afsar M, Khanam T, Kumar N, Ali F, Kumar S, Jahan F, Ramachandran R
|
| RgGuinier |
5.8 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
496 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transposon Tn3 resolvase dimer, 43 kDa Escherichia coli protein
Tn3 res, resolvase binding site II monomer, 32 kDa DNA
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 10 mM MgCl2, 1% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Nov 6
|
Structural basis for topological regulation of Tn3 resolvase.
Nucleic Acids Res (2022)
Montaño SP, Rowland SJ, Fuller JR, Burke ME, MacDonald AI, Boocock MR, Stark WM, Rice PA
|
| RgGuinier |
5.0 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA dC->dU-editing enzyme APOBEC-3G tetramer, 186 kDa Homo sapiens protein
40-mer single stranded inhibitory DNA dimer, 24 kDa DNA
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 6
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
395 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
40-mer single stranded inhibitory DNA monomer, 12 kDa DNA
DNA dC->dU-editing enzyme APOBEC-3G monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 6
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
|
|
|
|
|
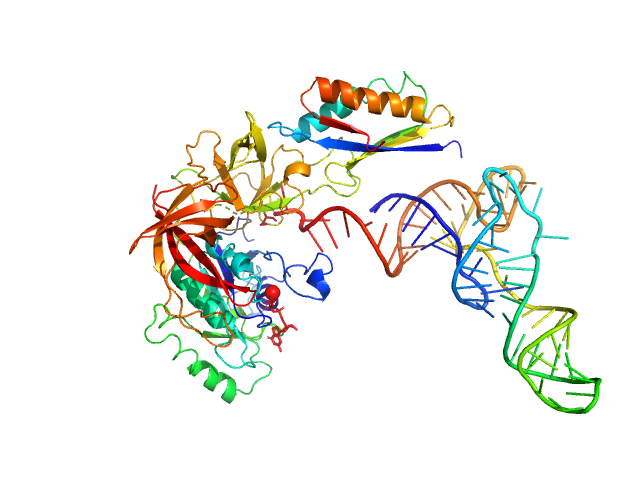
| Sample: |
Translation initiation factor 2 subunit gamma monomer, 46 kDa Saccharolobus solfataricus (strain … protein
Translation initiation factor 2 subunit alpha monomer, 10 kDa Saccharolobus solfataricus (strain … protein
Transfer RNA monomer, 23 kDa Escherichia coli RNA
|
| Buffer: |
10 mM MOPS- NaOH pH 6.7, 200 mM NaCl, 5 mM MgCl 2, 1 mM GDPNP, pH: 6.7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Nov 26
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
Gabel F, Engilberge S, Schmitt E, Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Homo sapiens RNA component of 7SK nuclear ribonucleoprotein (RN7SK), small nuclear RNA monomer, 18 kDa Homo sapiens RNA
Protein Tat monomer, 2 kDa Human immunodeficiency virus … protein
|
| Buffer: |
10 mM phosphate, 70 mM NaCl, 0.1 mM EDTA, pH: 5.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 27
|
A structure-based mechanism for displacement of the HEXIM adapter from 7SK small nuclear RNA.
Commun Biol 5(1):819 (2022)
Pham VV, Gao M, Meagher JL, Smith JL, D'Souza VM
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Homo sapiens RNA component of 7SK nuclear ribonucleoprotein (RN7SK), small nuclear RNA monomer, 18 kDa Homo sapiens RNA
Protein HEXIM1 monomer, 2 kDa Homo sapiens protein
|
| Buffer: |
10 mM phosphate, 70 mM NaCl, 0.1 mM EDTA, pH: 5.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 27
|
A structure-based mechanism for displacement of the HEXIM adapter from 7SK small nuclear RNA.
Commun Biol 5(1):819 (2022)
Pham VV, Gao M, Meagher JL, Smith JL, D'Souza VM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein Tat monomer, 2 kDa Human immunodeficiency virus … protein
Homo sapiens RNA component of 7SK nuclear ribonucleoprotein (RN7SK), small nuclear RNA monomer, 18 kDa Homo sapiens RNA
|
| Buffer: |
10 mM phosphate, 70 mM NaCl, 0.1 mM EDTA, pH: 5.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 9
|
A structure-based mechanism for displacement of the HEXIM adapter from 7SK small nuclear RNA.
Commun Biol 5(1):819 (2022)
Pham VV, Gao M, Meagher JL, Smith JL, D'Souza VM
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
28 |
nm3 |
|
|