|
|
|
|
|
| Sample: |
6S RNA (SsrS gene) monomer, 59 kDa Escherichia coli RNA
|
| Buffer: |
20 mM Tris-HCl, 200 mM KCl, 5 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 4
|
Probing the conformational changes of in vivo overexpressed cell cycle regulator 6S ncRNA
Frontiers in Molecular Biosciences 10 (2023)
Makraki E, Miliara S, Pagkalos M, Kokkinidis M, Mylonas E, Fadouloglou V
|
| RgGuinier |
6.1 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
6S RNA (SsrS gene) monomer, 59 kDa Escherichia coli RNA
Product RNA from E. coli 6S monomer, 7 kDa Escherichia coli RNA
|
| Buffer: |
20 mM Tris-HCl, 200 mM KCl, 5 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 4
|
Probing the conformational changes of in vivo overexpressed cell cycle regulator 6S ncRNA
Frontiers in Molecular Biosciences 10 (2023)
Makraki E, Miliara S, Pagkalos M, Kokkinidis M, Mylonas E, Fadouloglou V
|
| RgGuinier |
4.9 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQP8_ensemble_example.png)
|
| Sample: |
Stress response regulating small RNA OxyS monomer, 36 kDa Escherichia coli RNA
|
| Buffer: |
50 mM HEPES pH 6.8, 50 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at Austrian SAXS beamline 5.2L, ELETTRA on 2022 Jan 31
|
Spatial arrangement of functional domains in OxyS stress response sRNA
RNA :rna.079618.123 (2023)
Stih V, Amenitsch H, Plavec J, Podbevsek P
|
| RgGuinier |
4.8 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
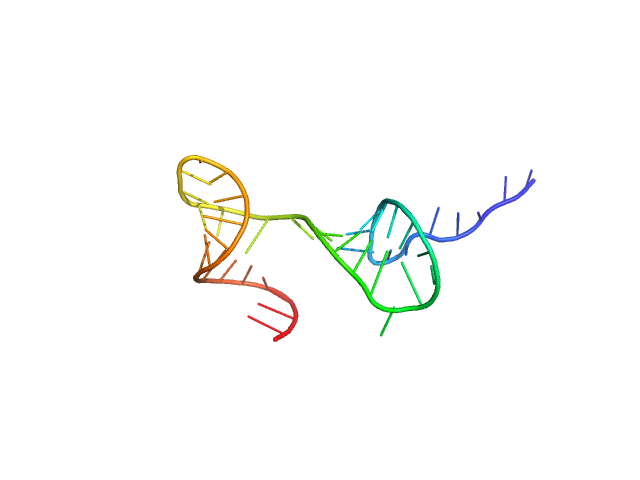
|
| Sample: |
Stem loop 2 and 3 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
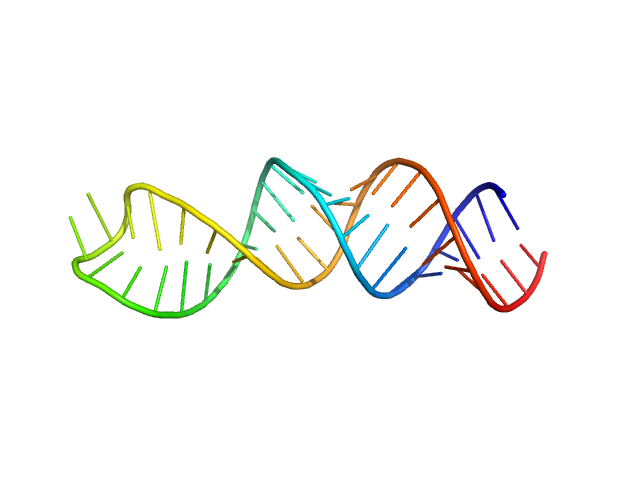
|
| Sample: |
Stem loop 4 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Stem loop 4 with AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 7 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Stem loop 2 and 3 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Stem loop 4 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Stem loop 4 with AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
35 |
nm3 |
|
|