|
|
|
|
|
| Sample: |
Truncated P5abc subdomain from tetrahymena ribozyme monomer, 18 kDa RNA
|
| Buffer: |
20mM KCl 0.25mM MgCl2 10mM KMOPS 20uM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Jun 17
|
Revealing the distinct folding phases of an RNA three-helix junction.
Nucleic Acids Res 46(14):7354-7365 (2018)
Plumridge A, Katz AM, Calvey GD, Elber R, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.6 |
nm |
|
|
|
|
|
|
|
| Sample: |
Truncated P5abc subdomain from tetrahymena ribozyme monomer, 18 kDa RNA
|
| Buffer: |
0.5mM MgCl2 20mM KCl 10mM KMOPS 20uM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Jun 17
|
Revealing the distinct folding phases of an RNA three-helix junction.
Nucleic Acids Res 46(14):7354-7365 (2018)
Plumridge A, Katz AM, Calvey GD, Elber R, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Truncated P5abc subdomain from tetrahymena ribozyme monomer, 18 kDa RNA
|
| Buffer: |
1mM MgCl2 20mM KCl 10mM KMOPS 20uM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Jun 17
|
Revealing the distinct folding phases of an RNA three-helix junction.
Nucleic Acids Res 46(14):7354-7365 (2018)
Plumridge A, Katz AM, Calvey GD, Elber R, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.2 |
nm |
|
|
|
|
|
|

|
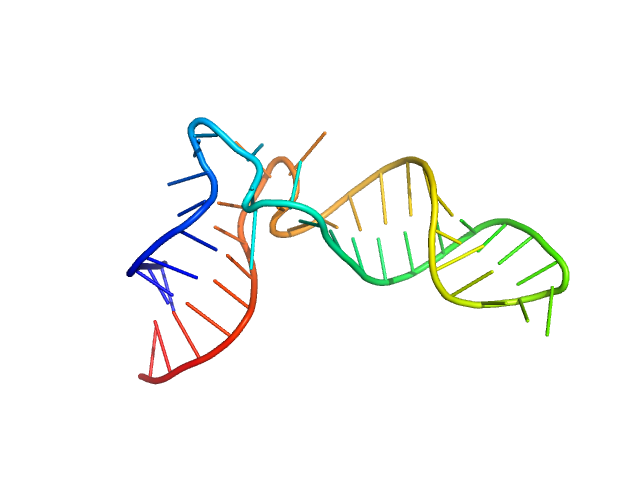
| Sample: |
Arginyl transfer RNA monomer, 13 kDa Romanomermis culicivorax RNA
|
| Buffer: |
50 mM HEPES-NaOH 10 mM MgCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Dec 16
|
Small but large enough: structural properties of armless mitochondrial tRNAs from the nematode Romanomermis culicivorax.
Nucleic Acids Res (2018)
Jühling T, Duchardt-Ferner E, Bonin S, Wöhnert J, Pütz J, Florentz C, Betat H, Sauter C, Mörl M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoleucyl transfer RNA monomer, 15 kDa Romanomermis culicivorax RNA
|
| Buffer: |
50 mM HEPES-NaOH 10 mM MgCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Dec 16
|
Small but large enough: structural properties of armless mitochondrial tRNAs from the nematode Romanomermis culicivorax.
Nucleic Acids Res (2018)
Jühling T, Duchardt-Ferner E, Bonin S, Wöhnert J, Pütz J, Florentz C, Betat H, Sauter C, Mörl M
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|

|
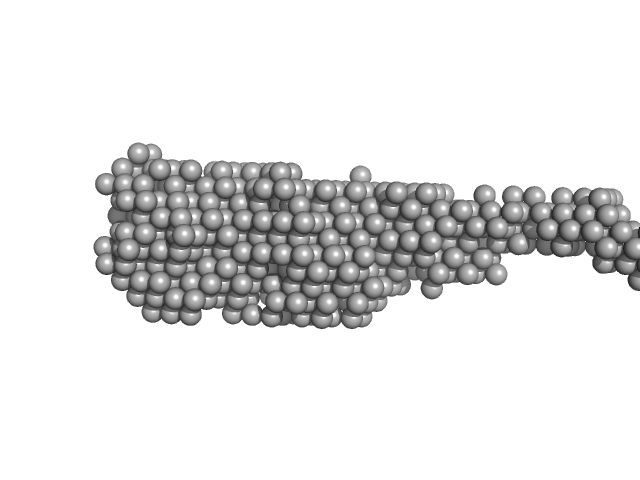
| Sample: |
Human telomerase RNA template component, DNA counterpart, nucleotides 1-20 dimer, 13 kDa Homo sapiens DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl,, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge.
Nucleic Acids Res 46(10):5319-5331 (2018)
Meier M, Moya-Torres A, Krahn NJ, McDougall MD, Orriss GL, McRae EKS, Booy EP, McEleney K, Patel TR, McKenna SA, Stetefeld J
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
E14A monomer, 21 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
E14B monomer, 13 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
1.9 |
nm |
| Dmax |
9.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
E14C monomer, 5 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
1.3 |
nm |
| Dmax |
5.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
E14AB monomer, 93 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
5.4 |
nm |
| Dmax |
24.7 |
nm |
|
|