|
|
|
|

|
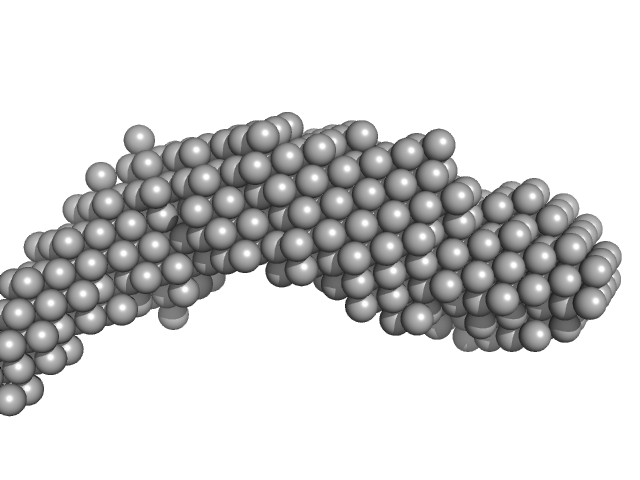
| Sample: |
E14ABC monomer, 106 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
4.7 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
E13A monomer, 17 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E13B monomer, 10 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E13C monomer, 6 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
1.3 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|
|
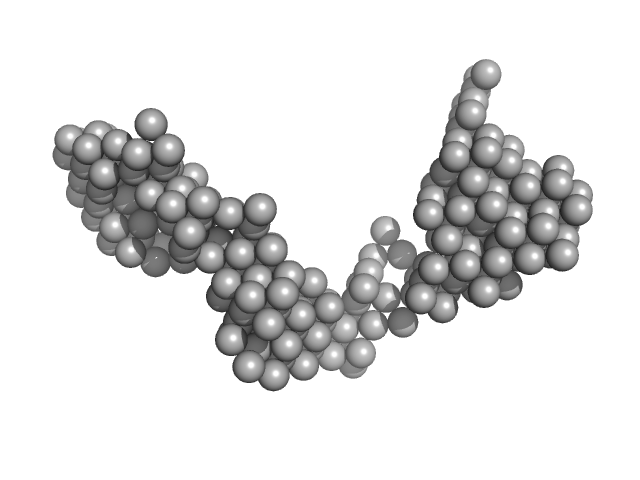
| Sample: |
E13AB monomer, 79 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
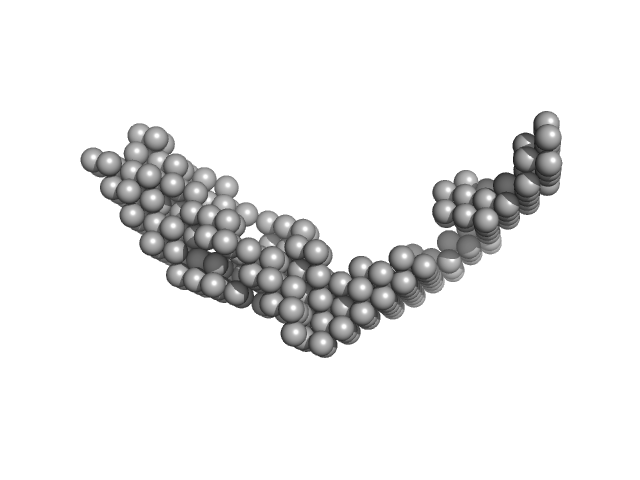
| Sample: |
E13ABC monomer, 42 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
3.9 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E13Ae14Be13C monomer, 40 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E14Ae13Be14C monomer, 31 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
3.4 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Methylated C-terminal ZBTB38 binding sequence monomer, 17 kDa DNA
|
| Buffer: |
10 mM Tris, 1 mM tris(2-carboxy-ethyl)phosphine (TCEP), 0.05% NaN3, 10% D2O, pH: 6.8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, Department of Chemistry, University of Utah on 2016 Oct 27
|
The C-Terminal Zinc Fingers of ZBTB38 are Novel Selective Readers of DNA Methylation.
J Mol Biol 430(3):258-271 (2018)
Pozner A, Hudson NO, Trewhella J, Terooatea TW, Miller SA, Buck-Koehntop BA
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Telomere DNA duplex monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 50 mM LiCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 May 3
|
Basic domain of telomere guardian TRF2 reduces D-loop unwinding whereas Rap1 restores it.
Nucleic Acids Res 45(21):12170-12180 (2017)
Necasová I, Janoušková E, Klumpler T, Hofr C
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
12 |
nm3 |
|
|