|
|
|
|
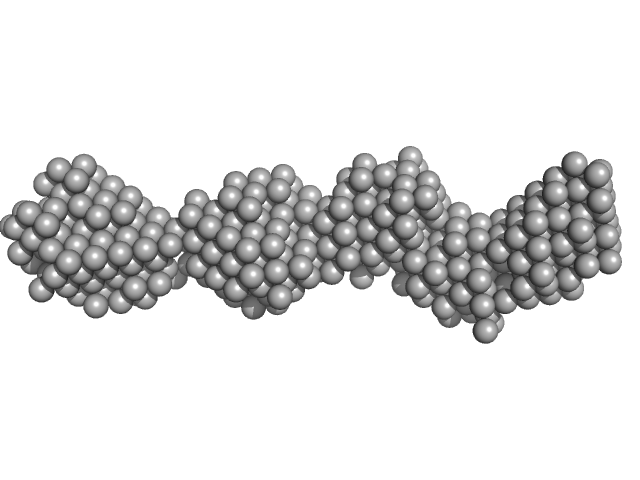
|
| Sample: |
40bp long dsDNA-Sa Oligonucleotide monomer, 25 kDa DNA
|
| Buffer: |
0.5 x Tris/Borate/EDTA (TBE), pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 28
|
Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius.
Mol Microbiol 105(5):777-793 (2017)
Li L, Banerjee A, Bischof LF, Maklad HR, Hoffmann L, Henche AL, Veliz F, Bildl W, Schulte U, Orell A, Essen LO, Peeters E, Albers SV
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|
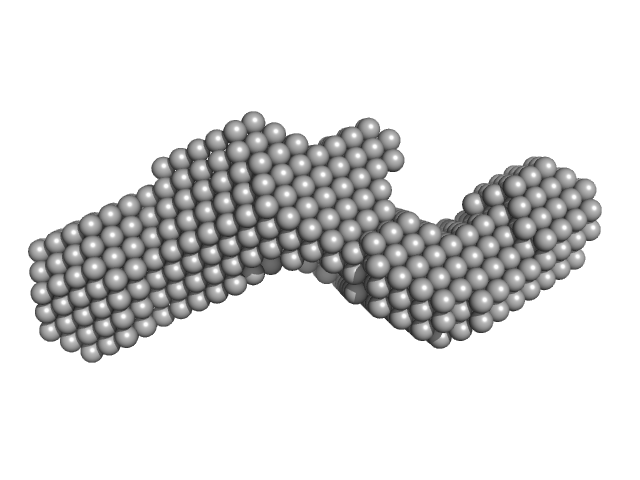
|
| Sample: |
Subdomain SL1 of hepatitis C virus monomer, 15 kDa Hepatitis C virus RNA
|
| Buffer: |
10mM Tris 0.1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 11
|
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.
RNA 23(9):1465-1476 (2017)
Cantero-Camacho Á, Fan L, Wang YX, Gallego J
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.1 |
nm |
|
|
|
|
|
|
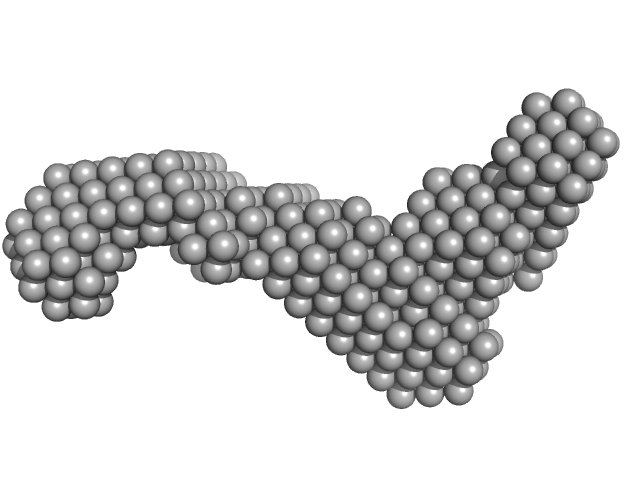
|
| Sample: |
Subdomain SL2' of hepatitis C virus domain 3'X monomer, 18 kDa Hepatitis C virus RNA
|
| Buffer: |
10mM Tris 0.1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 11
|
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.
RNA 23(9):1465-1476 (2017)
Cantero-Camacho Á, Fan L, Wang YX, Gallego J
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.0 |
nm |
|
|
|
|
|
|
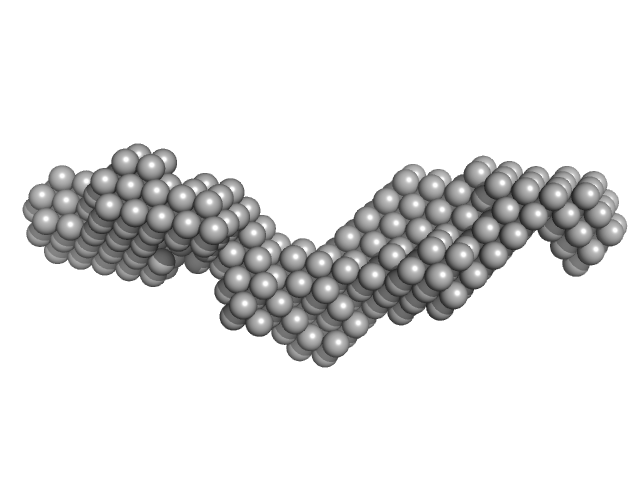
|
| Sample: |
Domain 3'X of hepatitis C virus monomer, 32 kDa Hepatitis C virus RNA
|
| Buffer: |
10mM Tris 0.1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 11
|
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.
RNA 23(9):1465-1476 (2017)
Cantero-Camacho Á, Fan L, Wang YX, Gallego J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.8 |
nm |
|
|
|
|
|
|

|
| Sample: |
Domain 3'X of hepatitis C virus dimer, 63 kDa Hepatitis C virus RNA
|
| Buffer: |
10mM Tris 0.1 mM EDTA 2 mM MgCl2 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Center for Cancer Research, National Cancer Institute on 2015 Nov 20
|
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.
RNA 23(9):1465-1476 (2017)
Cantero-Camacho Á, Fan L, Wang YX, Gallego J
|
| RgGuinier |
6.0 |
nm |
| Dmax |
24.4 |
nm |
|
|
|
|
|
|
|
| Sample: |
Subdomain SL2' of hepatitis C virus domain 3'X dimer, 36 kDa Hepatitis C virus RNA
|
| Buffer: |
10mM Tris 0,1mM EDTA 2mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 11
|
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.
RNA 23(9):1465-1476 (2017)
Cantero-Camacho Á, Fan L, Wang YX, Gallego J
|
| RgGuinier |
4.1 |
nm |
| Dmax |
16.9 |
nm |
|
|
|
|
|
|
|
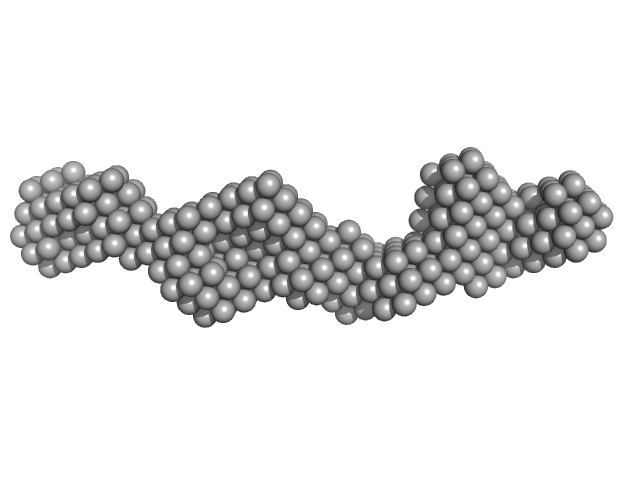
| Sample: |
Immediate Early 3 dimer, 12 kDa DNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Nov 12
|
The herpes viral transcription factor ICP4 forms a novel DNA recognition complex.
Nucleic Acids Res 45(13):8064-8078 (2017)
Tunnicliffe RB, Lockhart-Cairns MP, Levy C, Mould AP, Jowitt TA, Sito H, Baldock C, Sandri-Goldin RM, Golovanov AP
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.9 |
nm |
|
|
|
|
|
|
|
| Sample: |
Poly-deoxythymidine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Apr 1
|
Visualizing single-stranded nucleic acids in solution.
Nucleic Acids Res 45(9):e66 (2017)
Plumridge A, Meisburger SP, Pollack L
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.7 |
nm |
|
|
|
|
|
|
|
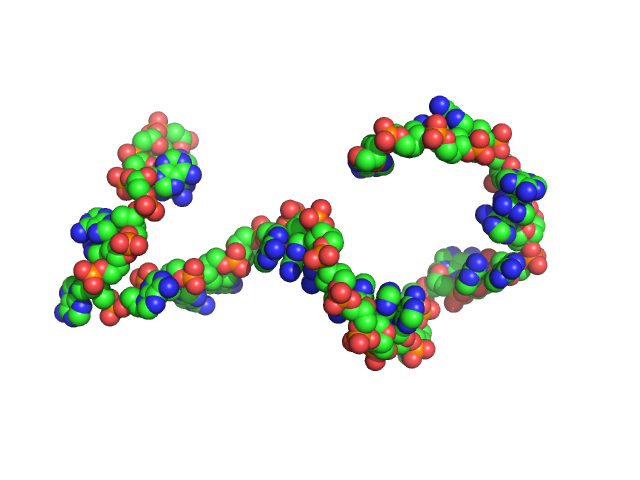
| Sample: |
Poly-deoxyadenosine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
Visualizing single-stranded nucleic acids in solution.
Nucleic Acids Res 45(9):e66 (2017)
Plumridge A, Meisburger SP, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Poly-deoxyadenosine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 100mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
The impact of base stacking on the conformations and electrostatics of single-stranded DNA.
Nucleic Acids Res 45(7):3932-3943 (2017)
Plumridge A, Meisburger SP, Andresen K, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
16 |
nm3 |
|
|