|
|
|
|
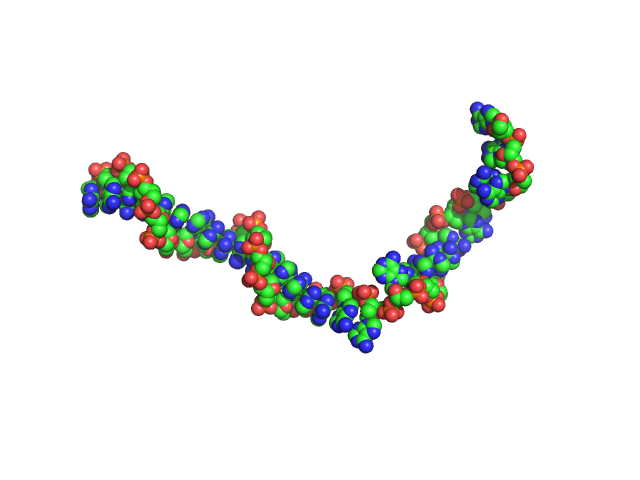
|
| Sample: |
Poly-deoxyadenosine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 200mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
The impact of base stacking on the conformations and electrostatics of single-stranded DNA.
Nucleic Acids Res 45(7):3932-3943 (2017)
Plumridge A, Meisburger SP, Andresen K, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
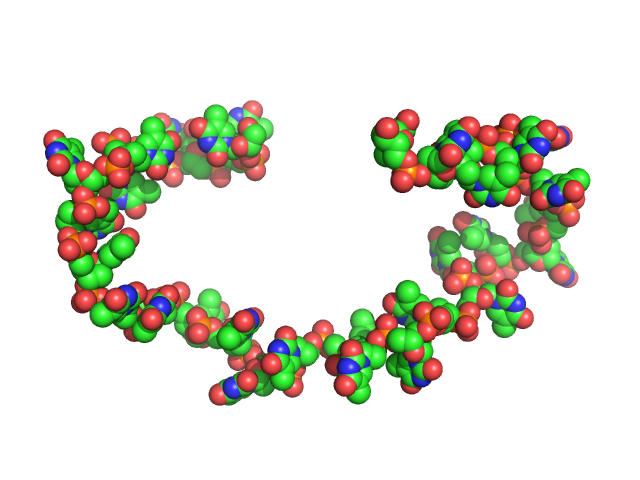
|
| Sample: |
Poly-deoxythymidine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 100mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
The impact of base stacking on the conformations and electrostatics of single-stranded DNA.
Nucleic Acids Res 45(7):3932-3943 (2017)
Plumridge A, Meisburger SP, Andresen K, Pollack L
|
|
|
|
|
|
|
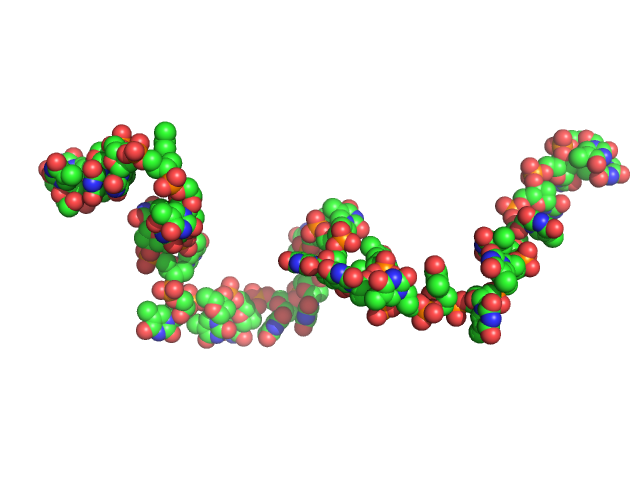
|
| Sample: |
Poly-deoxythymidine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 200mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
The impact of base stacking on the conformations and electrostatics of single-stranded DNA.
Nucleic Acids Res 45(7):3932-3943 (2017)
Plumridge A, Meisburger SP, Andresen K, Pollack L
|
|
|
|
|
|
|
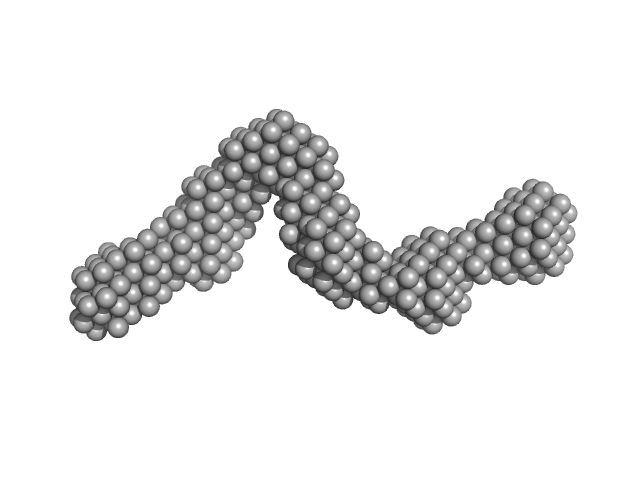
|
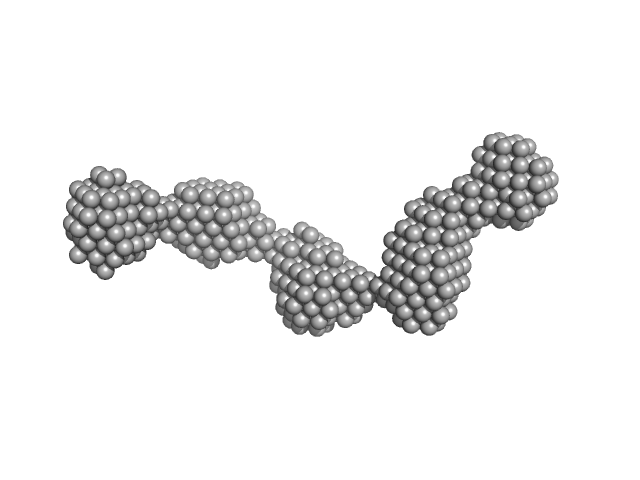
| Sample: |
AIR3-5FU dimer, 69 kDa RNA
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
4.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
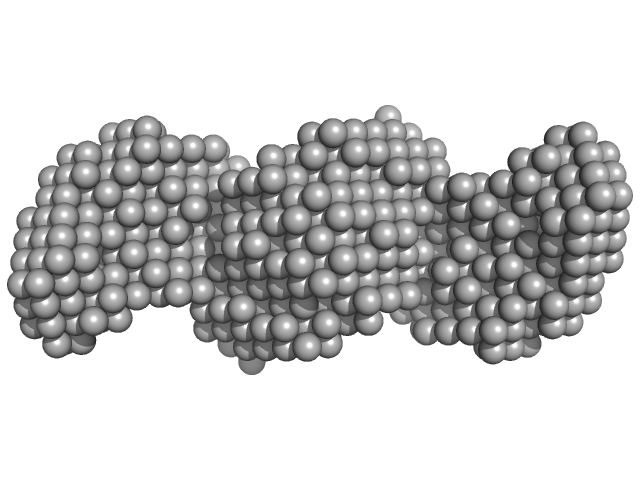
| Sample: |
AIR3-FdU dimer, 69 kDa RNA
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
AIR-3A dimer, 13 kDa RNA
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
AIR-3A 2'FU dimer, 13 kDa RNA
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
AIR-3A G18U tetramer, 25 kDa RNA
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|

|
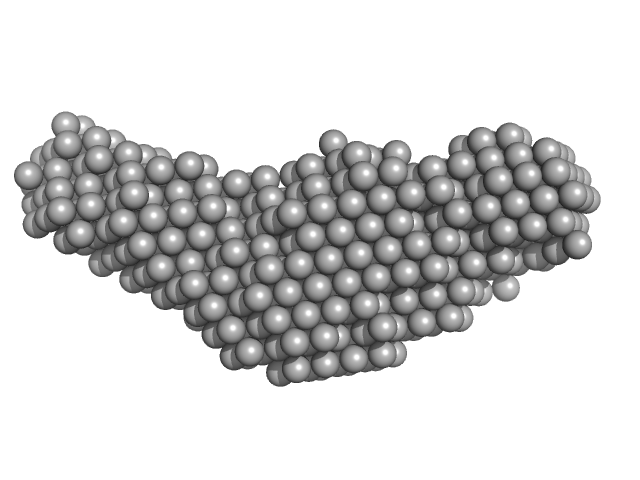
| Sample: |
DNA 22mer monomer, 12 kDa DNA
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 5% glycerol 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
The flexibility of a homeodomain transcription factor heterodimer and its allosteric regulation by DNA binding.
FEBS J 283(16):3134-54 (2016)
Mathiasen L, Valentini E, Boivin S, Cattaneo A, Blasi F, Svergun DI, Bruckmann C
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aureobox dsDNA monomer, 13 kDa synthetic construct DNA
|
| Buffer: |
50 mM Tris 50 mM boric acid 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 6
|
Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes.
Nucleic Acids Res 44(12):5957-70 (2016)
Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kroth PG, Essen LO, Kottke T
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
18 |
nm3 |
|
|