|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (588-1027: W988A mutant; Reverse transcriptase p51; p51 RT) monomer, 53 kDa Human immunodeficiency virus … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Jun 19
|
Biophysical Characterization of p51 and p66 Monomers of HIV-1 Reverse Transcriptase with Their Inhibitors.
Protein J (2023)
Seetaha S, Kamonsutthipaijit N, Yagi-Utsumi M, Seako Y, Yamaguchi T, Hannongbua S, Kato K, Choowongkomon K
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (588-1027: W988A mutant; Reverse transcriptase p51; p51 RT) monomer, 53 kDa Human immunodeficiency virus … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Jun 19
|
Biophysical Characterization of p51 and p66 Monomers of HIV-1 Reverse Transcriptase with Their Inhibitors.
Protein J (2023)
Seetaha S, Kamonsutthipaijit N, Yagi-Utsumi M, Seako Y, Yamaguchi T, Hannongbua S, Kato K, Choowongkomon K
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (588-1027: W988A mutant; Reverse transcriptase p51; p51 RT) monomer, 53 kDa Human immunodeficiency virus … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Jun 19
|
Biophysical Characterization of p51 and p66 Monomers of HIV-1 Reverse Transcriptase with Their Inhibitors.
Protein J (2023)
Seetaha S, Kamonsutthipaijit N, Yagi-Utsumi M, Seako Y, Yamaguchi T, Hannongbua S, Kato K, Choowongkomon K
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
226 |
nm3 |
|
|
|
|
|
|
|
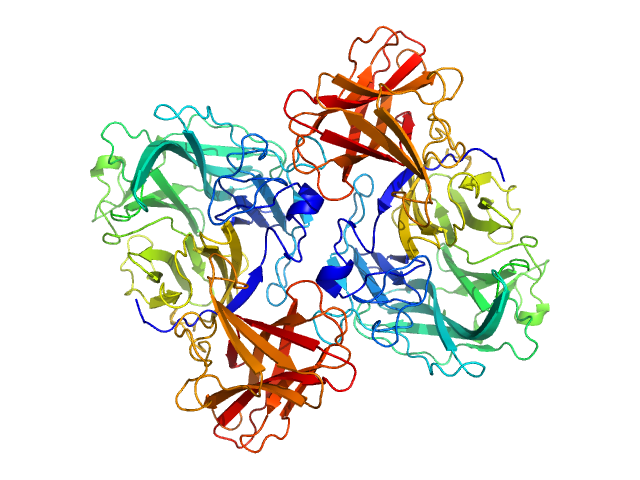
| Sample: |
Endo-D-arabinanase dimer, 107 kDa Microbacterium arabinogalactanolyticum protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Nov 3
|
Identification and characterization of endo-α-, exo-α-, and exo-β-d-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria
Nature Communications 14(1) (2023)
Shimokawa M, Ishiwata A, Kashima T, Nakashima C, Li J, Fukushima R, Sawai N, Nakamori M, Tanaka Y, Kudo A, Morikami S, Iwanaga N, Akai G, Shimizu N, Arakawa T, Yamada C, Kitahara K, Tanaka K, Ito Y, Fushinobu S, Fujita K
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
131 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
De novo enterobactin esterase Syn-F4 (K4T) dimer, 25 kDa synthetic construct protein
|
| Buffer: |
25 mM MES, 100 mM NaCl, 10% glycerol, 200 mM Arg-HCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Feb 11
|
Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc Natl Acad Sci U S A 120(38):e2218281120 (2023)
Kurihara K, Umezawa K, Donnelly AE, Sperling B, Liao G, Hecht MH, Arai R
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
|
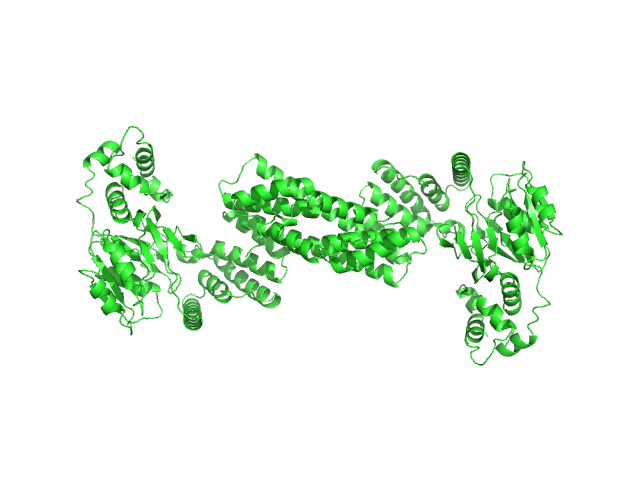
| Sample: |
Glyco_trans_2-like domain-containing protein dimer, 96 kDa Streptomyces sp. M41(2017) protein
|
| Buffer: |
20 mM Tris, pH 7.5, 100 mM NaCl, and 2 mM DTT + 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 18
|
Global shape of SvGT, a metal-dependent bacteriocin modifying S/O-HexNActransferase from actinobacteria: c -terminal dimerization modulates the function of this GT.
J Biomol Struct Dyn :1-15 (2023)
Sharma Y, Ahlawat S, Ashish, Rao A
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
|
|
|
|
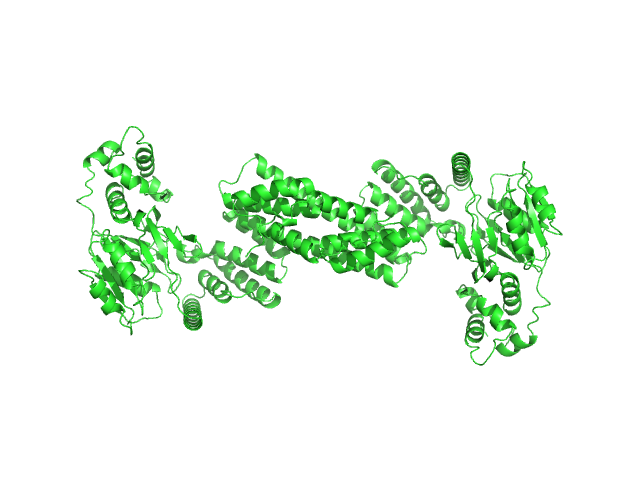
|
| Sample: |
Glyco_trans_2-like domain-containing protein dimer, 96 kDa Streptomyces sp. M41(2017) protein
|
| Buffer: |
20 mM Tris, pH 7.5, 100 mM NaCl, 2 mM DTT, 1 mM EDTA, 2 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 18
|
Global shape of SvGT, a metal-dependent bacteriocin modifying S/O-HexNActransferase from actinobacteria: c -terminal dimerization modulates the function of this GT.
J Biomol Struct Dyn :1-15 (2023)
Sharma Y, Ahlawat S, Ashish, Rao A
|
| RgGuinier |
5.8 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Major prion protein monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 13
|
A tetracationic porphyrin with dual anti-prion activity.
iScience 26(9):107480 (2023)
Masone A, Zucchelli C, Caruso E, Lavigna G, Eraña H, Giachin G, Tapella L, Comerio L, Restelli E, Raimondi I, Elezgarai SR, De Leo F, Quilici G, Taiarol L, Oldrati M, Lorenzo NL, García-Martínez S, Cagnotto A, Lucchetti J, Gobbi M, Vanni I, Nonno R, Di Bari MA, Tully MD, Cecatiello V, Ciossani G, Pasqualato S, Van Anken E, Salmona M, Castilla J, Requena JR, Banfi S, Musco G, Chiesa R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Major prion protein monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 13
|
A tetracationic porphyrin with dual anti-prion activity.
iScience 26(9):107480 (2023)
Masone A, Zucchelli C, Caruso E, Lavigna G, Eraña H, Giachin G, Tapella L, Comerio L, Restelli E, Raimondi I, Elezgarai SR, De Leo F, Quilici G, Taiarol L, Oldrati M, Lorenzo NL, García-Martínez S, Cagnotto A, Lucchetti J, Gobbi M, Vanni I, Nonno R, Di Bari MA, Tully MD, Cecatiello V, Ciossani G, Pasqualato S, Van Anken E, Salmona M, Castilla J, Requena JR, Banfi S, Musco G, Chiesa R
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
Major prion protein monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 13
|
A tetracationic porphyrin with dual anti-prion activity.
iScience 26(9):107480 (2023)
Masone A, Zucchelli C, Caruso E, Lavigna G, Eraña H, Giachin G, Tapella L, Comerio L, Restelli E, Raimondi I, Elezgarai SR, De Leo F, Quilici G, Taiarol L, Oldrati M, Lorenzo NL, García-Martínez S, Cagnotto A, Lucchetti J, Gobbi M, Vanni I, Nonno R, Di Bari MA, Tully MD, Cecatiello V, Ciossani G, Pasqualato S, Van Anken E, Salmona M, Castilla J, Requena JR, Banfi S, Musco G, Chiesa R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.3 |
nm |
|
|