|
|
|
|
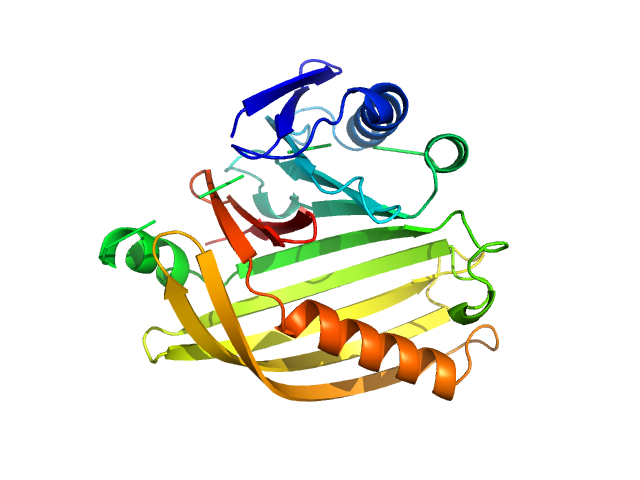
|
| Sample: |
Cholera toxin transcriptional activator monomer, 12 kDa Vibrio cholerae serotype … protein
Transmembrane regulatory protein ToxS dimer, 37 kDa Vibrio cholerae serotype … protein
Bile acid: sodium cholate hydrate monomer, 0 kDa
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, 3% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Jul 7
|
Vibrio cholerae's ToxRS bile sensing system.
Elife 12 (2023)
Gubensäk N, Sagmeister T, Buhlheller C, Geronimo BD, Wagner GE, Petrowitsch L, Gräwert MA, Rotzinger M, Berger TMI, Schäfer J, Usón I, Reidl J, Sánchez-Murcia PA, Zangger K, Pavkov-Keller T
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ubiquilin-2 (∆379-462) monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 8
|
Short disordered N-termini & proline-rich domain are major regulators of UBQLN1/2/4 phase separation.
Sarasi Galagedera
|
| RgGuinier |
4.4 |
nm |
| Dmax |
22.4 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ubiquilin-2 (∆487-538) dimer, 122 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 8
|
Short disordered N-termini & proline-rich domain are major regulators of UBQLN1/2/4 phase separation.
Sarasi Galagedera
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
288 |
nm3 |
|
|
|
|
|
|
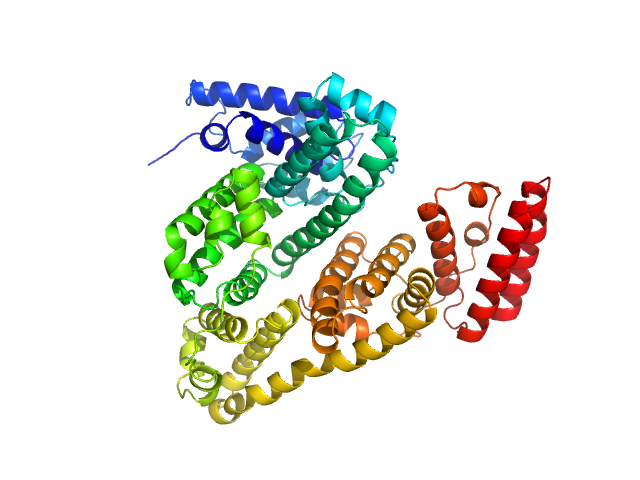
|
| Sample: |
Albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
10 mM HEPES, 5 mM NaCl, 0.1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Jun 8
|
Quantitative size-resolved characterization of mRNA nanoparticles by in-line coupling of asymmetrical-flow field-flow fractionation with small angle X-ray scattering.
Sci Rep 13(1):15764 (2023)
Graewert MA, Wilhelmy C, Bacic T, Schumacher J, Blanchet C, Meier F, Drexel R, Welz R, Kolb B, Bartels K, Nawroth T, Klein T, Svergun D, Langguth P, Haas H
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (588-1147: Reverse transcriptase/ribonuclease H; p66) monomer, 67 kDa Human immunodeficiency virus … protein
Gag-Pol polyprotein (588-1027: Reverse transcriptase p51; p51 RT) monomer, 53 kDa Human immunodeficiency virus … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Jun 19
|
Biophysical Characterization of p51 and p66 Monomers of HIV-1 Reverse Transcriptase with Their Inhibitors.
Protein J (2023)
Seetaha S, Kamonsutthipaijit N, Yagi-Utsumi M, Seako Y, Yamaguchi T, Hannongbua S, Kato K, Choowongkomon K
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
153 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (588-1147: Reverse transcriptase/ribonuclease H; p66) monomer, 67 kDa Human immunodeficiency virus … protein
Gag-Pol polyprotein (588-1027: Reverse transcriptase p51; p51 RT) monomer, 53 kDa Human immunodeficiency virus … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Jun 19
|
Biophysical Characterization of p51 and p66 Monomers of HIV-1 Reverse Transcriptase with Their Inhibitors.
Protein J (2023)
Seetaha S, Kamonsutthipaijit N, Yagi-Utsumi M, Seako Y, Yamaguchi T, Hannongbua S, Kato K, Choowongkomon K
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (588-1147: W988A mutant; Reverse transcriptase/ribonuclease H; p66) monomer, 67 kDa Human immunodeficiency virus … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Jun 19
|
Biophysical Characterization of p51 and p66 Monomers of HIV-1 Reverse Transcriptase with Their Inhibitors.
Protein J (2023)
Seetaha S, Kamonsutthipaijit N, Yagi-Utsumi M, Seako Y, Yamaguchi T, Hannongbua S, Kato K, Choowongkomon K
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (588-1147: Reverse transcriptase/ribonuclease H; p66) monomer, 67 kDa Human immunodeficiency virus … protein
Gag-Pol polyprotein (588-1027: Reverse transcriptase p51; p51 RT) monomer, 53 kDa Human immunodeficiency virus … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Jun 19
|
Biophysical Characterization of p51 and p66 Monomers of HIV-1 Reverse Transcriptase with Their Inhibitors.
Protein J (2023)
Seetaha S, Kamonsutthipaijit N, Yagi-Utsumi M, Seako Y, Yamaguchi T, Hannongbua S, Kato K, Choowongkomon K
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
178 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (588-1147: W988A mutant; Reverse transcriptase/ribonuclease H; p66) monomer, 67 kDa Human immunodeficiency virus … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Jun 19
|
Biophysical Characterization of p51 and p66 Monomers of HIV-1 Reverse Transcriptase with Their Inhibitors.
Protein J (2023)
Seetaha S, Kamonsutthipaijit N, Yagi-Utsumi M, Seako Y, Yamaguchi T, Hannongbua S, Kato K, Choowongkomon K
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gag-Pol polyprotein (588-1147: W988A mutant; Reverse transcriptase/ribonuclease H; p66) monomer, 67 kDa Human immunodeficiency virus … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Jun 19
|
Biophysical Characterization of p51 and p66 Monomers of HIV-1 Reverse Transcriptase with Their Inhibitors.
Protein J (2023)
Seetaha S, Kamonsutthipaijit N, Yagi-Utsumi M, Seako Y, Yamaguchi T, Hannongbua S, Kato K, Choowongkomon K
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
270 |
nm3 |
|
|