|
|
|
|
|
| Sample: |
Endo-beta-N-acetylglucosaminidase H dimer, 61 kDa Streptomyces plicatus protein
|
| Buffer: |
20 mM Tris-HCl, 50 mM NaCl, 5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Nov 4
|
SAXS studies of X-ray induced disulfide bond damage: Engineering high-resolution insight from a low-resolution technique
PLOS ONE 15(11):e0239702 (2020)
Stachowski T, Snell M, Snell E, Boggon T
|
| RgGuinier |
2.7 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endo-beta-N-acetylglucosaminidase H dimer, 61 kDa Streptomyces plicatus protein
|
| Buffer: |
20 mM Tris-HCl, 50 mM NaCl, 5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Nov 4
|
SAXS studies of X-ray induced disulfide bond damage: Engineering high-resolution insight from a low-resolution technique
PLOS ONE 15(11):e0239702 (2020)
Stachowski T, Snell M, Snell E, Boggon T
|
| RgGuinier |
2.7 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endo-beta-N-acetylglucosaminidase H dimer, 61 kDa Streptomyces plicatus protein
|
| Buffer: |
20 mM Tris-HCl, 50 mM NaCl, 5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Nov 4
|
SAXS studies of X-ray induced disulfide bond damage: Engineering high-resolution insight from a low-resolution technique
PLOS ONE 15(11):e0239702 (2020)
Stachowski T, Snell M, Snell E, Boggon T
|
| RgGuinier |
2.8 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endo-beta-N-acetylglucosaminidase H dimer, 61 kDa Streptomyces plicatus protein
|
| Buffer: |
20 mM Tris-HCl, 50 mM NaCl, 5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Nov 4
|
SAXS studies of X-ray induced disulfide bond damage: Engineering high-resolution insight from a low-resolution technique
PLOS ONE 15(11):e0239702 (2020)
Stachowski T, Snell M, Snell E, Boggon T
|
| RgGuinier |
2.7 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoprotein monomer, 45 kDa Nipah henipavirus protein
|
| Buffer: |
20 mM Tris-HCl, 0.3 M NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 3
|
Ensemble description of the intrinsically disordered N-terminal domain of the Nipah virus P/V protein from combined NMR and SAXS.
Sci Rep 10(1):19574 (2020)
Schiavina M, Salladini E, Murrali MG, Tria G, Felli IC, Pierattelli R, Longhi S
|
| RgGuinier |
6.2 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Synthetic nanobody Sybody 23 monomer, 16 kDa synthetic construct protein
|
| Buffer: |
50 mM Tris 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 May 5
|
Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2
Nature Communications 11(1) (2020)
Custódio T, Das H, Sheward D, Hanke L, Pazicky S, Pieprzyk J, Sorgenfrei M, Schroer M, Gruzinov A, Jeffries C, Graewert M, Svergun D, Dobrev N, Remans K, Seeger M, McInerney G, Murrell B, Hällberg B, Löw C
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Spike glycoprotein (ACE2 receptor binding domain) monomer, 29 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM Tris 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 May 1
|
Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2
Nature Communications 11(1) (2020)
Custódio T, Das H, Sheward D, Hanke L, Pazicky S, Pieprzyk J, Sorgenfrei M, Schroer M, Gruzinov A, Jeffries C, Graewert M, Svergun D, Dobrev N, Remans K, Seeger M, McInerney G, Murrell B, Hällberg B, Löw C
|
| RgGuinier |
3.0 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
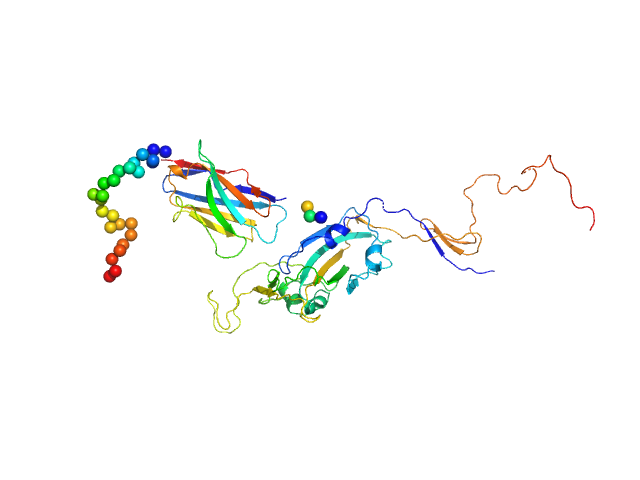
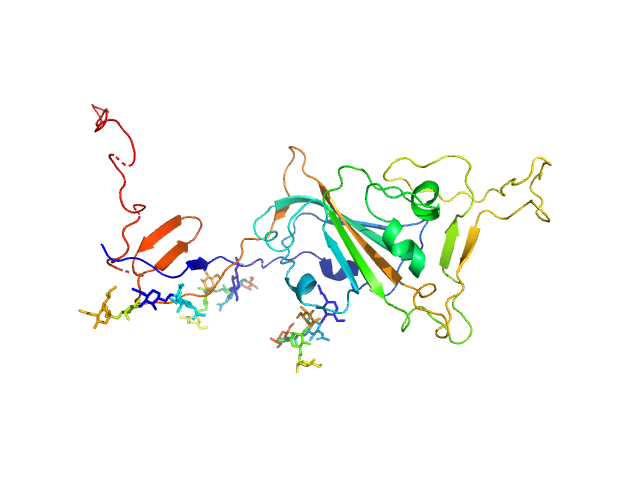
|
| Sample: |
Synthetic nanobody Sybody 23 monomer, 16 kDa synthetic construct protein
Spike glycoprotein (ACE2 receptor binding domain) monomer, 29 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM Tris 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 May 10
|
Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2
Nature Communications 11(1) (2020)
Custódio T, Das H, Sheward D, Hanke L, Pazicky S, Pieprzyk J, Sorgenfrei M, Schroer M, Gruzinov A, Jeffries C, Graewert M, Svergun D, Dobrev N, Remans K, Seeger M, McInerney G, Murrell B, Hällberg B, Löw C
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Apoferritin light chain 24-mer, 479 kDa Equus caballus protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 5
|
Adding Size Exclusion Chromatography (SEC) and Light Scattering (LS) Devices to Obtain High-Quality Small Angle X-Ray Scattering (SAXS) Data
Crystals 10(11):975 (2020)
Graewert M, Da Vela S, Gräwert T, Molodenskiy D, Blanchet C, Svergun D, Jeffries C
|
| RgGuinier |
5.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
679 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Carbonic anhydrase 2 monomer, 29 kDa Bos taurus protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 5
|
Adding Size Exclusion Chromatography (SEC) and Light Scattering (LS) Devices to Obtain High-Quality Small Angle X-Ray Scattering (SAXS) Data
Crystals 10(11):975 (2020)
Graewert M, Da Vela S, Gräwert T, Molodenskiy D, Blanchet C, Svergun D, Jeffries C
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.1 |
nm |
| VolumePorod |
37 |
nm3 |
|
|