|
|
|
|
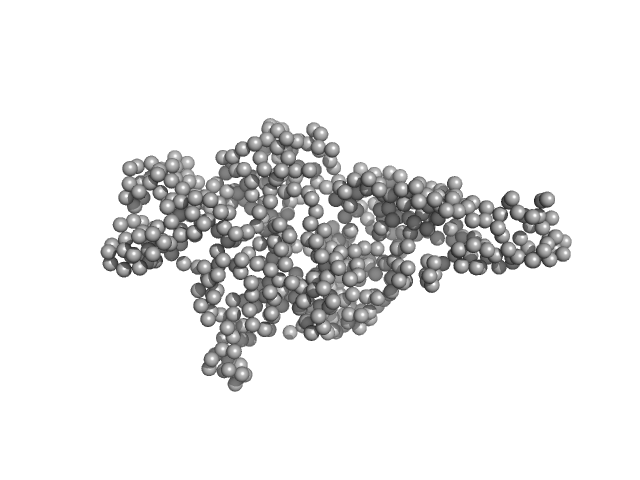
|
| Sample: |
Hepatocyte growth factor receptor monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 27
|
Dimerization of kringle 1 domain from hepatocyte growth factor/scatter factor provides a potent MET receptor agonist
Life Science Alliance 5(12):e202201424 (2022)
de Nola G, Leclercq B, Mougel A, Taront S, Simonneau C, Forneris F, Adriaenssens E, Drobecq H, Iamele L, Dubuquoy L, Melnyk O, Gherardi E, de Jonge H, Vicogne J
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
|
|
|
|
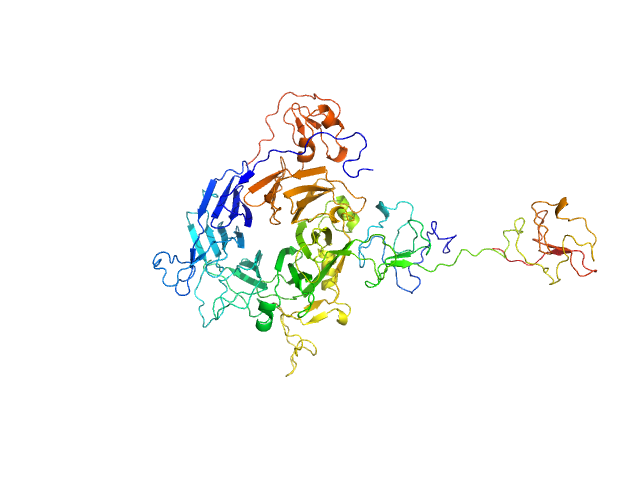
|
| Sample: |
Minimal hepatocyte growth factor mimic K1K1 monomer, 19 kDa synthetic construct protein
Hepatocyte growth factor receptor monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 23
|
Dimerization of kringle 1 domain from hepatocyte growth factor/scatter factor provides a potent MET receptor agonist
Life Science Alliance 5(12):e202201424 (2022)
de Nola G, Leclercq B, Mougel A, Taront S, Simonneau C, Forneris F, Adriaenssens E, Drobecq H, Iamele L, Dubuquoy L, Melnyk O, Gherardi E, de Jonge H, Vicogne J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
136 |
nm3 |
|
|
|
|
|
|
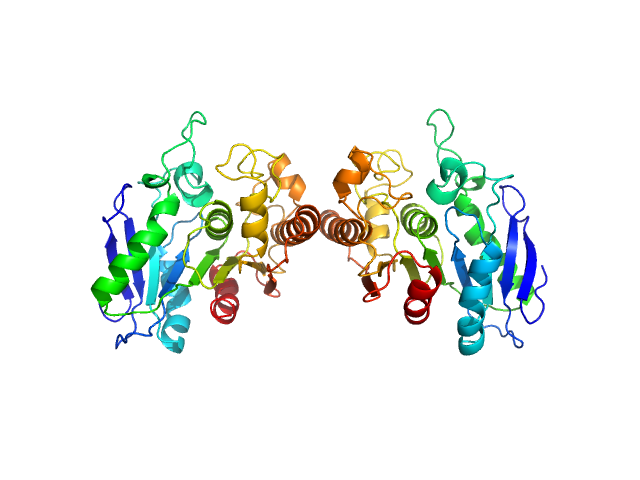
|
| Sample: |
Poly(Aspartic acid) hydrolase-1 dimer, 65 kDa Sphingomonas sp. KT-1 protein
|
| Buffer: |
20 mM Tris pH 7.0, 100 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 27
|
Structural Characterization of Sphingomonas
sp. KT-1 PahZ1-Catalyzed Biodegradation of Thermally Synthesized Poly(aspartic acid)
ACS Sustainable Chemistry & Engineering (2020)
Brambley C, Bolay A, Salvo H, Jansch A, Yared T, Miller J, Wallen J, Weiland M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Oct 19
|
The first DEP domain of the RhoGEF P-Rex1 autoinhibits activity andcontributes to membrane binding.
J Biol Chem (2020)
Ravala SK, Hopkins JB, Plescia CB, Allgood SR, Kane MA, Cash JN, Stahelin RV, Tesmer JJG
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Oct 19
|
The first DEP domain of the RhoGEF P-Rex1 autoinhibits activity andcontributes to membrane binding.
J Biol Chem (2020)
Ravala SK, Hopkins JB, Plescia CB, Allgood SR, Kane MA, Cash JN, Stahelin RV, Tesmer JJG
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Oct 19
|
The first DEP domain of the RhoGEF P-Rex1 autoinhibits activity andcontributes to membrane binding.
J Biol Chem (2020)
Ravala SK, Hopkins JB, Plescia CB, Allgood SR, Kane MA, Cash JN, Stahelin RV, Tesmer JJG
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Pentafunctional AROM polypeptide dimer, 345 kDa Chaetomium thermophilum protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl. 2 mM TCEP, 1% sucrose, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Nov 22
|
Architecture and functional dynamics of the pentafunctional AROM complex
Nature Chemical Biology (2020)
Arora Verasztó H, Logotheti M, Albrecht R, Leitner A, Zhu H, Hartmann M
|
| RgGuinier |
5.4 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
623 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Plasmodium falciparum Heat shock protein 90 dimer, 177 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM KCl, 1 mM β-mercaptoethanol, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 11
|
Solution structure of Plasmodium falciparum Hsp90 indicates a high flexible dimer
Archives of Biochemistry and Biophysics :108468 (2020)
Silva N, Torricillas M, Minari K, Barbosa L, Seraphim T, Borges J
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Plasmodium falciparum Heat shock protein 90 N-terminal and Middle domains monomer, 68 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM KCl, 1 mM β-mercaptoethanol, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 11
|
Solution structure of Plasmodium falciparum Hsp90 indicates a high flexible dimer
Archives of Biochemistry and Biophysics :108468 (2020)
Silva N, Torricillas M, Minari K, Barbosa L, Seraphim T, Borges J
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Plasmodium falciparum Heat shock protein 90 middle domain monomer, 33 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM KCl, 1 mM β-mercaptoethanol, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 11
|
Solution structure of Plasmodium falciparum Hsp90 indicates a high flexible dimer
Archives of Biochemistry and Biophysics :108468 (2020)
Silva N, Torricillas M, Minari K, Barbosa L, Seraphim T, Borges J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
47 |
nm3 |
|
|