|
|
|
|
|
| Sample: |
Adhesion G-protein coupled receptor G6 S2 D134A/F135A monomer, 86 kDa Danio rerio protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Jun 28
|
Structural basis for adhesion G protein-coupled receptor Gpr126 function
Nature Communications 11(1) (2020)
Leon K, Cunningham R, Riback J, Feldman E, Li J, Sosnick T, Zhao M, Monk K, Araç D
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Adhesion G-protein coupled receptor G6 S2 monomer, 88 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Jun 28
|
Structural basis for adhesion G protein-coupled receptor Gpr126 function
Nature Communications 11(1) (2020)
Leon K, Cunningham R, Riback J, Feldman E, Li J, Sosnick T, Zhao M, Monk K, Araç D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
199 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Adhesion G-protein coupled receptor G6 S1 monomer, 91 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Jun 28
|
Structural basis for adhesion G protein-coupled receptor Gpr126 function
Nature Communications 11(1) (2020)
Leon K, Cunningham R, Riback J, Feldman E, Li J, Sosnick T, Zhao M, Monk K, Araç D
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
|
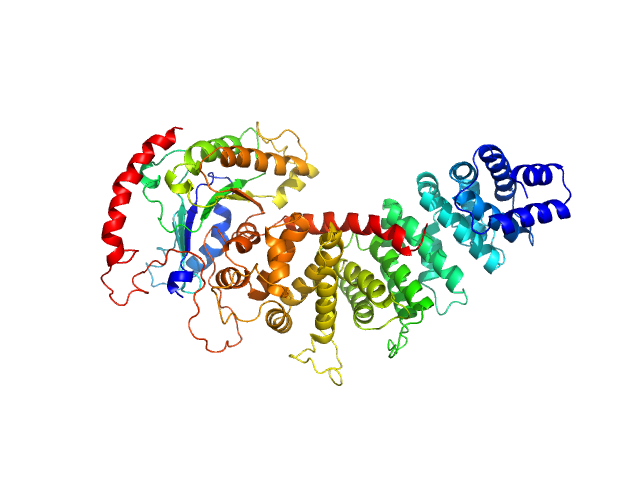
| Sample: |
Resistance to inhibitors of cholinesterase 8 homolog A monomer, 56 kDa Bos taurus protein
MiniGi monomer, 25 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 5 % glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Oct 27
|
Large-scale conformational rearrangement of the α5-helix of Gα subunits in complex with the guanine nucleotide exchange factor Ric8A.
J Biol Chem (2019)
Srivastava D, Artemyev NO
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
|
|
|
|
|
|
|
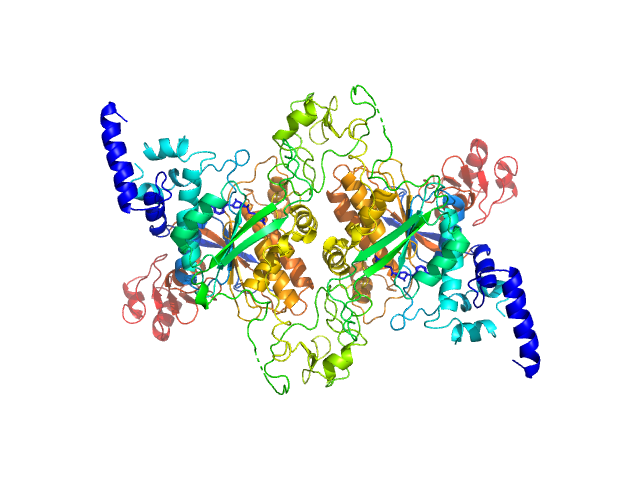
| Sample: |
Adenine specific DNA methyltransferase (Mod) dimer, 137 kDa Helicobacter pylori protein
|
| Buffer: |
25 mM Tris, 250 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2017 Jul 9
|
Tetramerization at low pH licenses DNA methylation activity of M.HpyAXI in the presence of acid stress.
J Mol Biol (2019)
Narayanan N, Banerjee A, Jain D, Kulkarni DS, Sharma R, Nirwal S, Rao DN, Nair DT
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
143 |
nm3 |
|
|
|
|
|
|
|
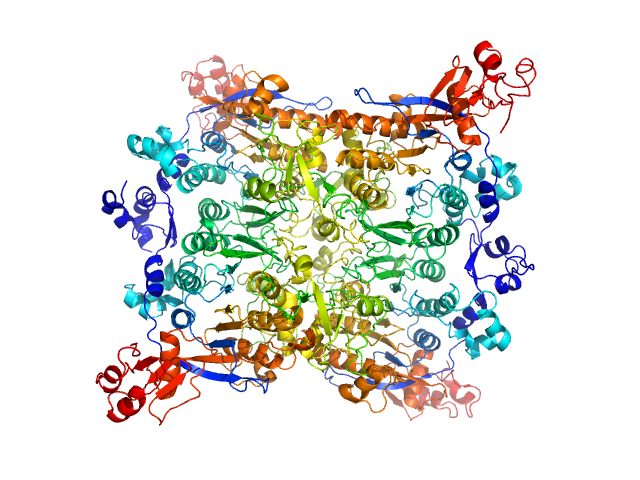
| Sample: |
Adenine specific DNA methyltransferase (Mod) tetramer, 273 kDa Helicobacter pylori protein
|
| Buffer: |
25 mM citrate, 250 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 1
|
Tetramerization at low pH licenses DNA methylation activity of M.HpyAXI in the presence of acid stress.
J Mol Biol (2019)
Narayanan N, Banerjee A, Jain D, Kulkarni DS, Sharma R, Nirwal S, Rao DN, Nair DT
|
| RgGuinier |
5.0 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
316 |
nm3 |
|
|
|
|
|
|
|
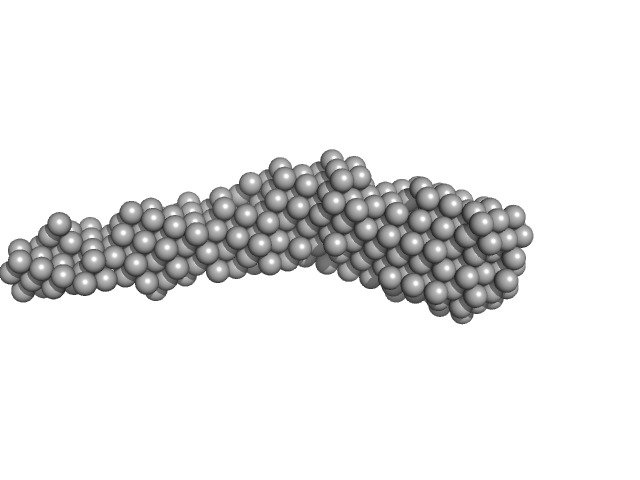
| Sample: |
Flagella binding tail protein monomer, 103 kDa Salmonella virus Chi protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.03 % NaN3, 5.0 % glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 4
|
The flagellotropic bacteriophage YSD1 targets Salmonella Typhi with a Chi-like protein tail fibre.
Mol Microbiol (2019)
Dunstan RA, Pickard D, Dougan S, Goulding D, Cormie C, Hardy J, Li F, Grinter R, Harcourt K, Yu L, Song J, Schreiber F, Choudhary J, Clare S, Coulibaly F, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
5.6 |
nm |
| Dmax |
27.4 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|
|
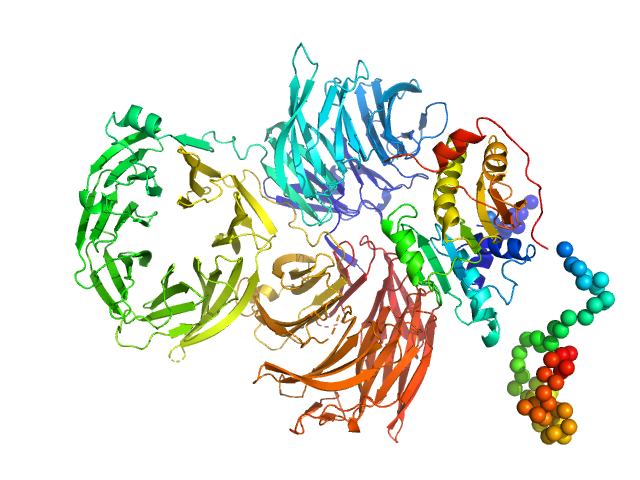
| Sample: |
Trm7: tRNA (cytidine(32)/guanosine(34)-2'-O)-methyltransferase monomer, 36 kDa Saccharomyces cerevisiae protein
Trm734: Regulator of Ty1 transposition protein 10 monomer, 116 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 5% v/v Glycerol, 10mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2015 Dec 16
|
Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition.
Nucleic Acids Res (2019)
Hirata A, Okada K, Yoshii K, Shiraishi H, Saijo S, Yonezawa K, Shimizu N, Hori H
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
|
|
|
|

|
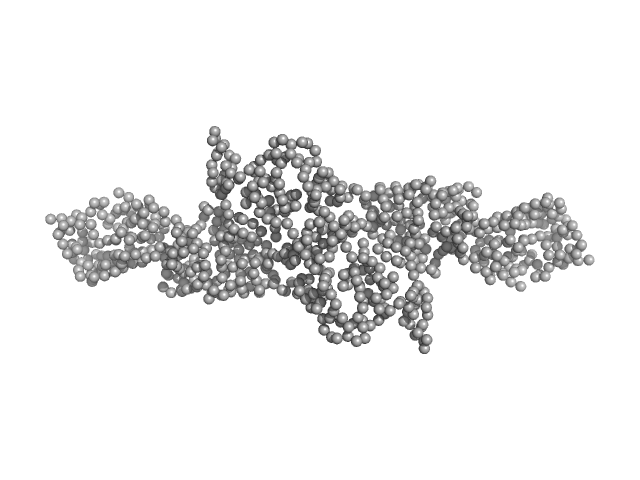
| Sample: |
Cytohesin-2; ARNO truncation mutant monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Nov 25
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cytohesin-2 ARF nucleotide-binding site opener dimer, 93 kDa Homo sapiens protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 23
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.7 |
nm |
| VolumePorod |
145 |
nm3 |
|
|