|
|
|
|
|
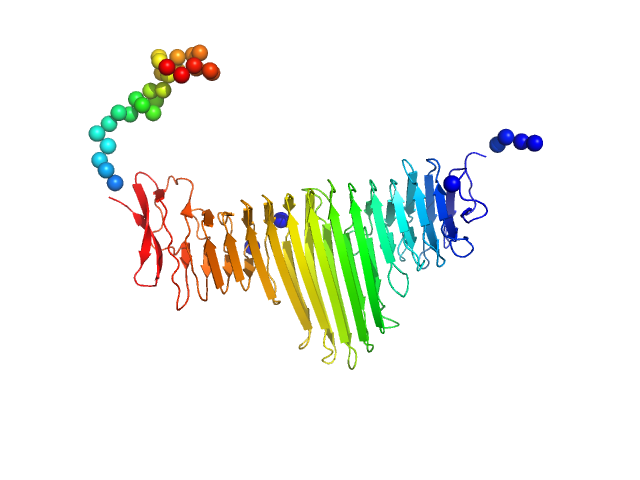
| Sample: |
Apolipoprotein E4 tetramer, 139 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 29
|
The molecular basis for Apolipoprotein E4 as the major risk factor for late onset Alzheimer's disease.
J Mol Biol (2019)
Raulin AC, Kraft L, Al-Hilaly YK, Xue WF, McGeehan JE, Atack JR, Serpell L
|
| RgGuinier |
5.8 |
nm |
| Dmax |
19.6 |
nm |
| VolumePorod |
430 |
nm3 |
|
|
|
|
|
|
|
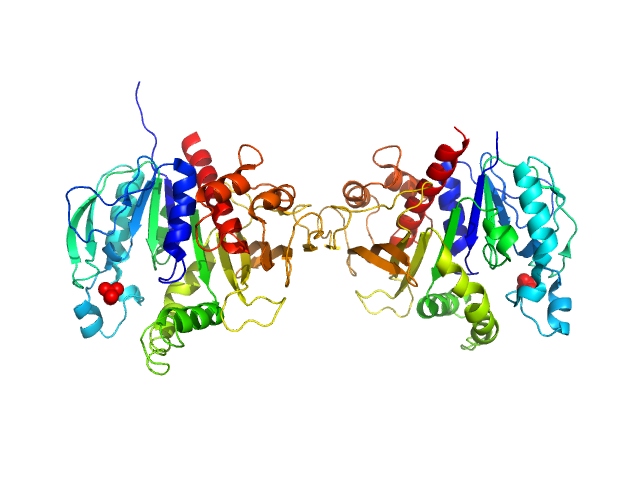
| Sample: |
Alpha domain of autotransporter protein UpaB monomer, 48 kDa E. Coli CFT073 protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 May 1
|
Unique structural features of a bacterial autotransporter adhesin suggest mechanisms for interaction with host macromolecules.
Nat Commun 10(1):1967 (2019)
Paxman JJ, Lo AW, Sullivan MJ, Panjikar S, Kuiper M, Whitten AE, Wang G, Luan CH, Moriel DG, Tan L, Peters KM, Phan MD, Gee CL, Ulett GC, Schembri MA, Heras B
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
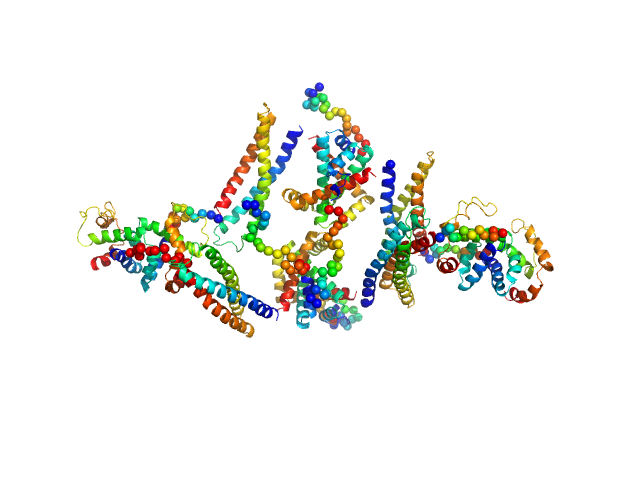
| Sample: |
Phosphoribulokinase, chloroplastic dimer, 78 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
Tris-HCl 50 mM 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 15
|
Arabidopsis and Chlamydomonas phosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc Natl Acad Sci U S A 116(16):8048-8053 (2019)
Gurrieri L, Del Giudice A, Demitri N, Falini G, Pavel NV, Zaffagnini M, Polentarutti M, Crozet P, Marchand CH, Henri J, Trost P, Lemaire SD, Sparla F, Fermani S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
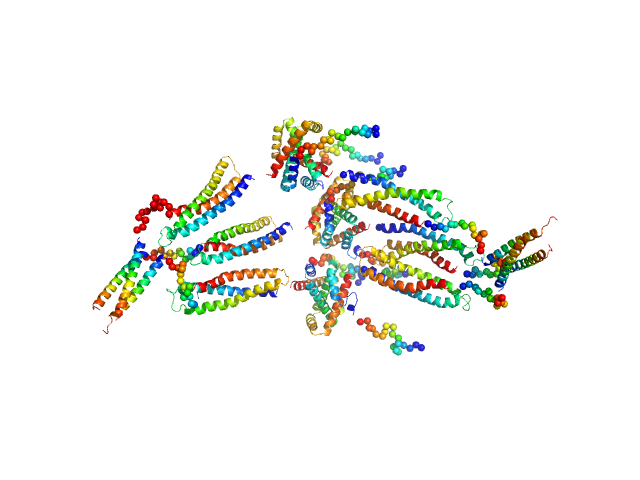
| Sample: |
HP2042 form from Helicobacter pylori, N-terminal domain of syntaxin-1A from Rattus norvegicus, Trp repressor from Escherichia coli tetramer, 150 kDa Helicobacter pylori, Rattus … protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku Nano-Viewer, Nara Institute of Science and Technology on 2016 Dec 8
|
Construction of a Quadrangular Tetramer and a Cage-Like Hexamer from Three-Helix Bundle-Linked Fusion Proteins.
ACS Synth Biol (2019)
Miyamoto T, Hayashi Y, Yoshida K, Watanabe H, Uchihashi T, Yonezawa K, Shimizu N, Kamikubo H, Hirota S
|
| RgGuinier |
5.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
379 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HP0242 from Helicobacter pylori, N-terminal domain of syntaxin-1A from Rattus norvegicus, de novo designed coiled-coil trimer domain hexamer, 179 kDa Helicobacter pylori, Rattus … protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku Nano-Viewer, Nara Institute of Science and Technology on 2017 Mar 29
|
Construction of a Quadrangular Tetramer and a Cage-Like Hexamer from Three-Helix Bundle-Linked Fusion Proteins.
ACS Synth Biol (2019)
Miyamoto T, Hayashi Y, Yoshida K, Watanabe H, Uchihashi T, Yonezawa K, Shimizu N, Kamikubo H, Hirota S
|
| RgGuinier |
6.5 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
642 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysyne-specific Demethylase LSD2 monomer, 89 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 200 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 11
|
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep 27(2):387-399.e7 (2019)
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
124 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NPAC dehydrogenase domain tetramer, 125 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 200 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 26
|
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep 27(2):387-399.e7 (2019)
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
167 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NPAC linker+DH (delta-205) tetramer, 150 kDa Homo sapiens protein
|
| Buffer: |
15 mM HEPES, 200 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 26
|
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep 27(2):387-399.e7 (2019)
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A
|
| RgGuinier |
4.0 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
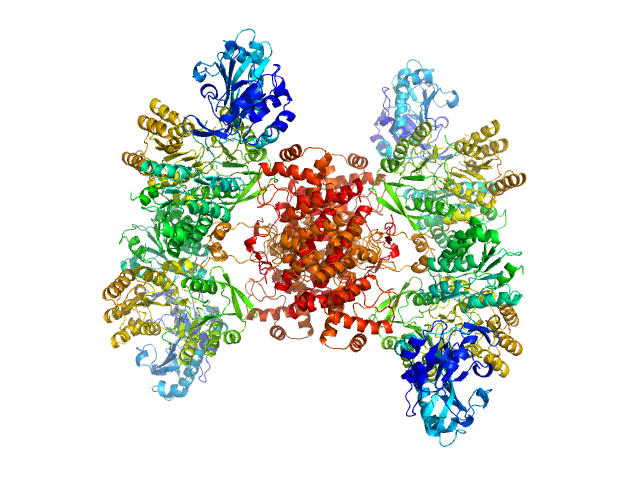
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 6
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.0 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
738 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 6
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
747 |
nm3 |
|
|