|
|
|
|
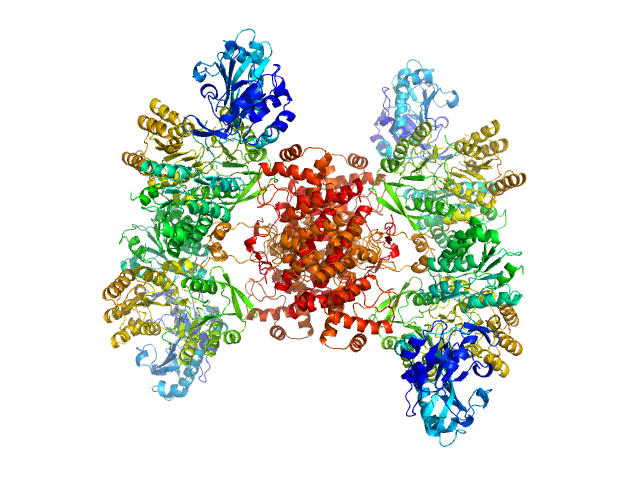
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 6
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.0 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
738 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 6
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
747 |
nm3 |
|
|
|
|
|
|
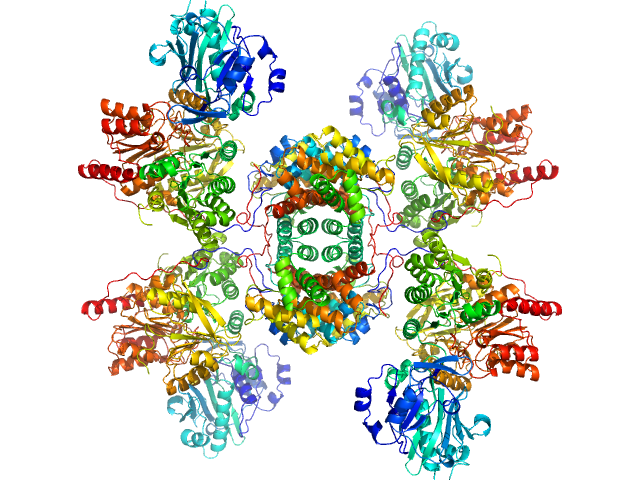
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, 2mM CoA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 5
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
5.8 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
709 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate lyase tetramer, 429 kDa Chlorobium limicola protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 4
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
666 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate lyase tetramer, 429 kDa Chlorobium limicola protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 4
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
672 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate lyase tetramer, 429 kDa Chlorobium limicola protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, 2mM CoA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 4
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
5.7 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
791 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 4
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.1 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
765 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 4
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.2 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
787 |
nm3 |
|
|
|
|
|
|
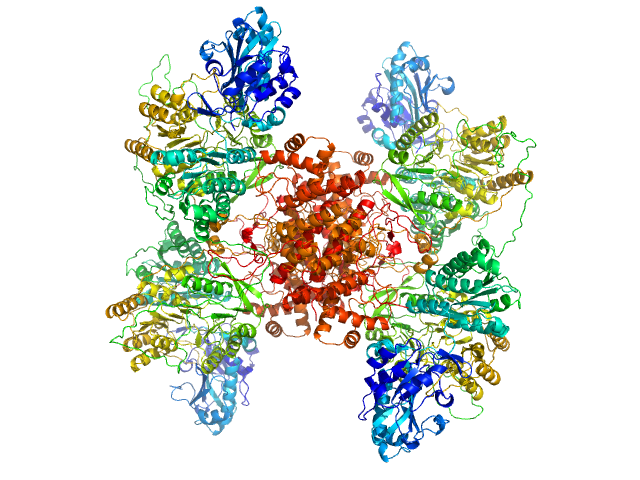
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, 2mM CoA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 4
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
5.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
775 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polyglutamine-binding protein 1 p.Lys192Serfs*7 dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
Phosphate-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2013 Feb 15
|
Frameshift PQBP-1 mutants K192Sfs*7 and R153Sfs*41 implicated in X-linked intellectual disability form stable dimers.
J Struct Biol (2019)
Rahman SK, Okazawa H, Chen YW
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
114 |
nm3 |
|
|