|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Urokinase-type plasminogen activator (Amino Terminal Fragment) monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, 50 mM NaSO4,, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 18
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 1
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Urokinase-type plasminogen activator (Amino Terminal Fragment) monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 1
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 5
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Urokinase-type plasminogen activator (Amino Terminal Fragment) monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 5
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cyclic GMP-AMP synthase monomer, 61 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 25
|
cGAS facilitates sensing of extracellular cyclic dinucleotides to activate innate immunity.
EMBO Rep (2019)
Liu H, Moura-Alves P, Pei G, Mollenkopf HJ, Hurwitz R, Wu X, Wang F, Liu S, Ma M, Fei Y, Zhu C, Koehler AB, Oberbeck-Mueller D, Hahnke K, Klemm M, Guhlich-Bornhof U, Ge B, Tuukkanen A, Kolbe M, Dorhoi A, Kaufmann SH
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cyclic GMP-AMP synthase dimer, 123 kDa Homo sapiens protein
2'-O,5'-O-((adenosine-3'-O,5'-O-diyl)bisphosphinico)guanosine dimer, 1 kDa
|
| Buffer: |
20 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 25
|
cGAS facilitates sensing of extracellular cyclic dinucleotides to activate innate immunity.
EMBO Rep (2019)
Liu H, Moura-Alves P, Pei G, Mollenkopf HJ, Hurwitz R, Wu X, Wang F, Liu S, Ma M, Fei Y, Zhu C, Koehler AB, Oberbeck-Mueller D, Hahnke K, Klemm M, Guhlich-Bornhof U, Ge B, Tuukkanen A, Kolbe M, Dorhoi A, Kaufmann SH
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|
|
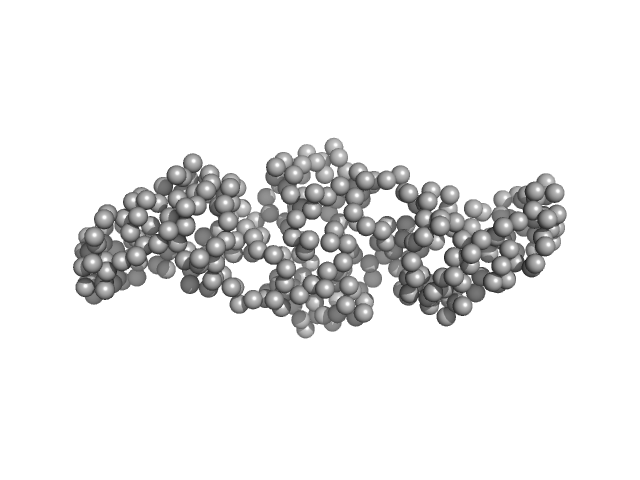
| Sample: |
Gliding motility protein MglB dimer, 34 kDa Myxococcus xanthus protein
|
| Buffer: |
150 mM NaCl, 1 mM DTT, 20 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 9
|
MglA functions as a three-state GTPase to control movement reversals of Myxococcus xanthus.
Nat Commun 10(1):5300 (2019)
Galicia C, Lhospice S, Varela PF, Trapani S, Zhang W, Navaza J, Herrou J, Mignot T, Cherfils J
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
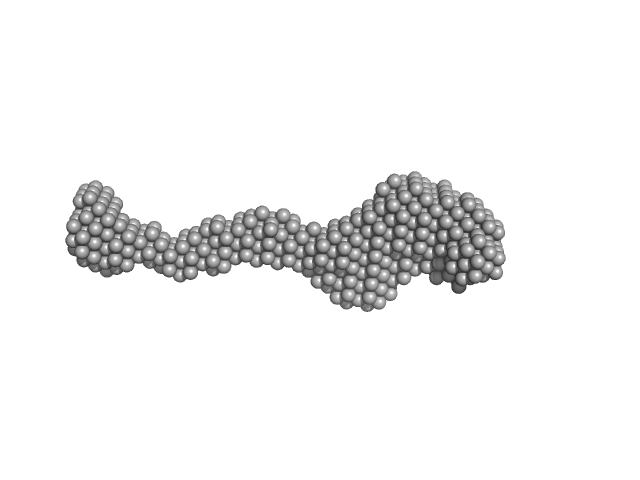
| Sample: |
Lysine-specific demethylase 5B monomer, 176 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 300 mM NaCl, 5% (v/v) glycerol, 1mM DTT, pH: 7.7 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with GeniX3D, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Oct 24
|
Molecular architecture of the Jumonji C family histone demethylase KDM5B.
Sci Rep 9(1):4019 (2019)
Dorosz J, Kristensen LH, Aduri NG, Mirza O, Lousen R, Bucciarelli S, Mehta V, Sellés-Baiget S, Solbak SMØ, Bach A, Mesa P, Hernandez PA, Montoya G, Nguyen TTTN, Rand KD, Boesen T, Gajhede M
|
| RgGuinier |
8.8 |
nm |
| Dmax |
26.9 |
nm |
|
|
|
|
|
|
|
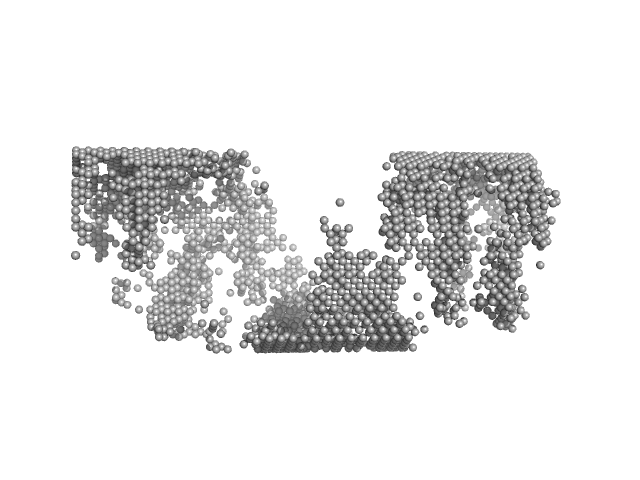
| Sample: |
Matrix protein, 40 kDa Newcastle disease virus … protein
|
| Buffer: |
STE buffer 100 mM NaCl, 10 mM Tris-HCl, and 1 mM EDTA, pH: 4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 11
|
Solution Structure, Self-Assembly, and Membrane Interactions of the Matrix Protein from Newcastle Disease Virus at Neutral and Acidic pH
Journal of Virology 93(6) (2019)
Shtykova E, Petoukhov M, Dadinova L, Fedorova N, Tashkin V, Timofeeva T, Ksenofontov A, Loshkarev N, Baratova L, Jeffries C, Svergun D, Batishchev O, García-Sastre A
|
|
|