|
|
|
|
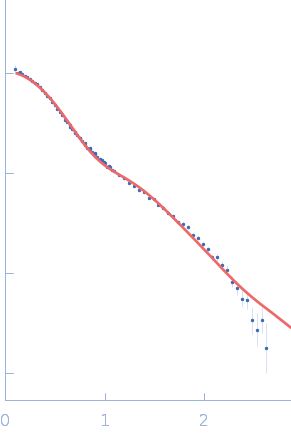
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 11
|
NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation unique to this diflavin reductase
Journal of Structural Biology 205(2):170-179 (2019)
Tavolieri A, Murray D, Askenasy I, Pennington J, McGarry L, Stanley C, Stroupe M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Diadenylate cyclase dimer, 39 kDa Staphylococcus aureus protein
Phosphoglucosamine mutase dimer, 99 kDa Staphylococcus aureus protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 7
|
Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM.
PLoS Pathog 15(1):e1007537 (2019)
Tosi T, Hoshiga F, Millership C, Singh R, Eldrid C, Patin D, Mengin-Lecreulx D, Thalassinos K, Freemont P, Gründling A
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Phosphoglucosamine mutase dimer, 99 kDa Staphylococcus aureus protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 7
|
Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM.
PLoS Pathog 15(1):e1007537 (2019)
Tosi T, Hoshiga F, Millership C, Singh R, Eldrid C, Patin D, Mengin-Lecreulx D, Thalassinos K, Freemont P, Gründling A
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Diadenylate cyclase dimer, 39 kDa Staphylococcus aureus protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 7
|
Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM.
PLoS Pathog 15(1):e1007537 (2019)
Tosi T, Hoshiga F, Millership C, Singh R, Eldrid C, Patin D, Mengin-Lecreulx D, Thalassinos K, Freemont P, Gründling A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
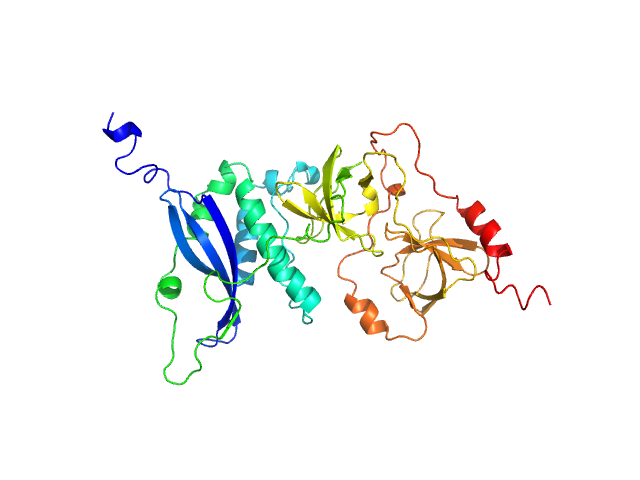
|
| Sample: |
Neutophil cytosol factor 1 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM EDTA, 2 mM DTT, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, IBBMC on 2009 Oct 16
|
Quantitative live-cell imaging and 3D modeling reveal critical functional features in the cytosolic complex of phagocyte NADPH oxidase.
J Biol Chem (2019)
Ziegler CS, Bouchab L, Tramier M, Durand D, Fieschi F, Dupré-Crochet S, Mérola F, Nüße O, Erard M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
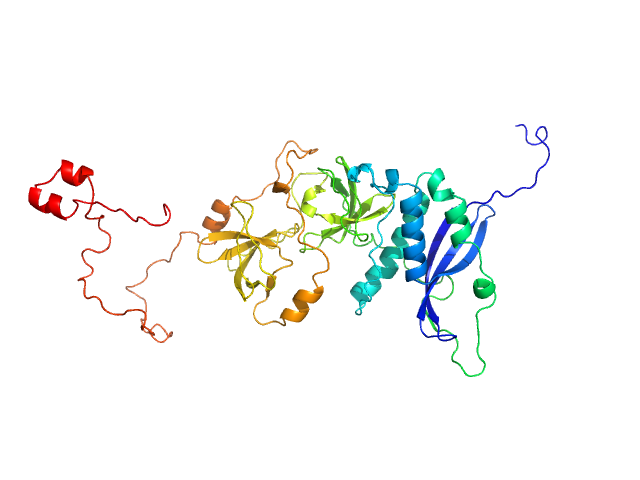
|
| Sample: |
Neutrophil cytosol factor 1 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM EDTA, 2 mM DTT, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2008 Apr 23
|
Quantitative live-cell imaging and 3D modeling reveal critical functional features in the cytosolic complex of phagocyte NADPH oxidase.
J Biol Chem (2019)
Ziegler CS, Bouchab L, Tramier M, Durand D, Fieschi F, Dupré-Crochet S, Mérola F, Nüße O, Erard M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neutrophil cytosol factor 2 monomer, 61 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 50 mM NaCl, 1 mM EDTA, 2 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at D24, LURE on 2003 Apr 9
|
Quantitative live-cell imaging and 3D modeling reveal critical functional features in the cytosolic complex of phagocyte NADPH oxidase.
J Biol Chem (2019)
Ziegler CS, Bouchab L, Tramier M, Durand D, Fieschi F, Dupré-Crochet S, Mérola F, Nüße O, Erard M
|
| RgGuinier |
4.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
|
|
|
|
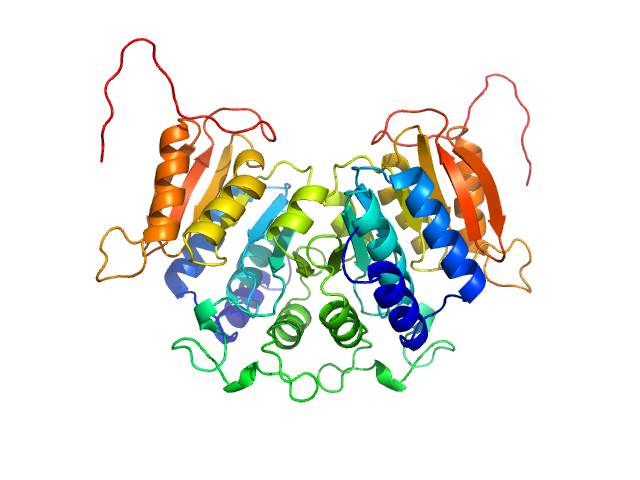
|
| Sample: |
Methyltransferase domain protein dimer, 51 kDa Mycolicibacterium hassiacum protein
|
| Buffer: |
20 mM bis-tris propane, 50 mM NaCl, 2 mM DTT, 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jun 21
|
Biosynthesis of mycobacterial methylmannose polysaccharides requires a unique 1-O-methyltransferase specific for 3-O-methylated mannosides.
Proc Natl Acad Sci U S A 116(3):835-844 (2019)
Ripoll-Rozada J, Costa M, Manso JA, Maranha A, Miranda V, Sequeira A, Ventura MR, Macedo-Ribeiro S, Pereira PJB, Empadinhas N
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
|
|
|
|
|
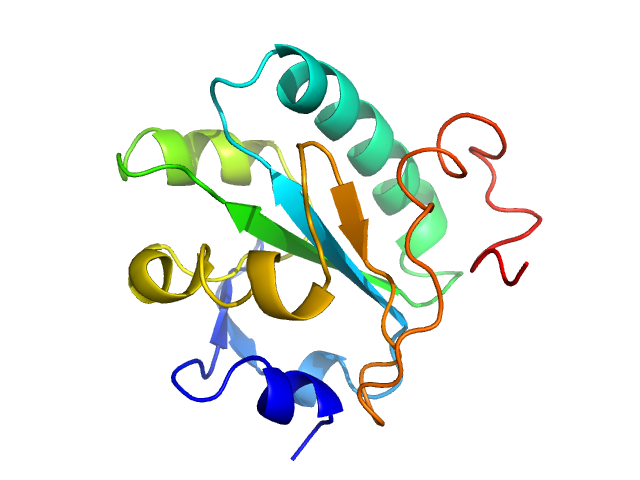
| Sample: |
Tryparedoxin monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Tryparedoxin monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
27 |
nm3 |
|
|