|
|
|
|
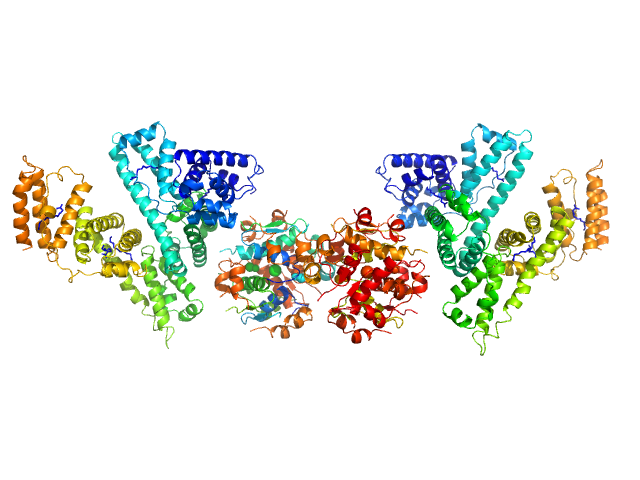
|
| Sample: |
Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.) monomer, 66 kDa protein
Insulin detemir (Levemir(R), Novo Nordisk A/S) dodecamer, 71 kDa protein
|
| Buffer: |
8.8 mM Na2HPO4, 10.6 mM m-cresol, 12.2 mM phenol, 140.9 mM glycerol, 56.9 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 Sep 25
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
309 |
nm3 |
|
|
|
|
|
|
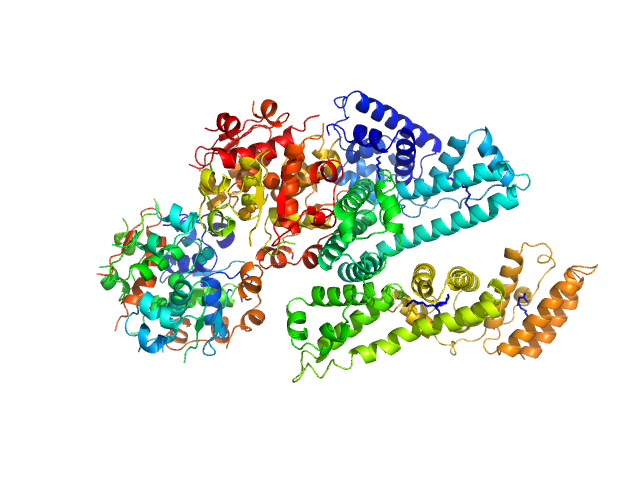
|
| Sample: |
Human Albumin (Recombumin(R) Elite, Albumedix Ltd.) monomer, 66 kDa protein
Insulin degludec(Tresiba(R), Novo Nordisk A/S) dodecamer, 73 kDa protein
|
| Buffer: |
25 mM Na2HPO4, 15.9 mM m-cresol, 15.9 mM phenol, 212.8 mM glycerol, 20 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 27
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
179 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Putative killer protein, Toxin GraT. monomer, 11 kDa Pseudomonas putida protein
|
| Buffer: |
50 mM Tris 250 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Sep 21
|
A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun 10(1):972 (2019)
Talavera A, Tamman H, Ainelo A, Konijnenberg A, Hadži S, Sobott F, Garcia-Pino A, Hõrak R, Loris R
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Putative killer protein, Toxin GraT. monomer, 11 kDa Pseudomonas putida protein
Antitoxin GraA, antidote of toxin GraT dimer, 22 kDa Pseudomonas putida protein
Putative killer protein, Toxin GraT monomer, 11 kDa Pseudomonas putida protein
|
| Buffer: |
50 mM Tris 250 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 29
|
A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun 10(1):972 (2019)
Talavera A, Tamman H, Ainelo A, Konijnenberg A, Hadži S, Sobott F, Garcia-Pino A, Hõrak R, Loris R
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
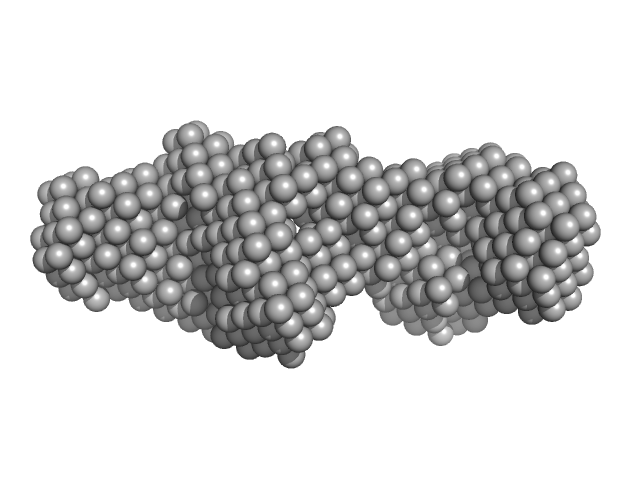
|
| Sample: |
LIM/homeobox protein Lhx4 monomer, 15 kDa Mus musculus protein
Insulin gene enhancer protein ISL-2 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins (2019)
Stokes PH, Robertson NO, Silva AP, Estephan T, Trewhella J, Guss JM, Matthews JM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
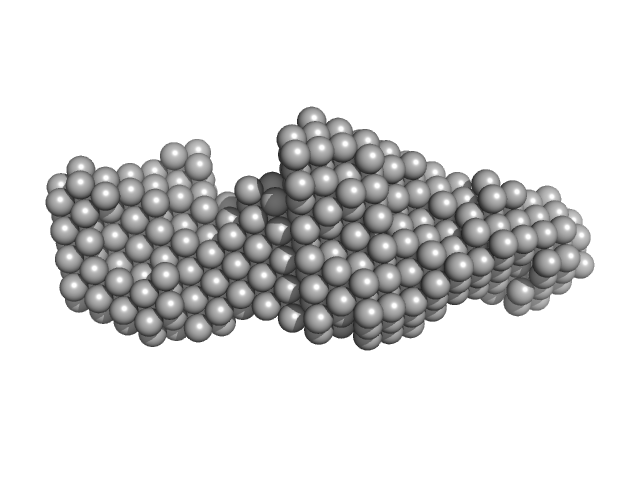
|
| Sample: |
LIM/homeobox protein Lhx4 monomer, 15 kDa Mus musculus protein
Insulin gene enhancer protein ISL-2 (R282G) monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins (2019)
Stokes PH, Robertson NO, Silva AP, Estephan T, Trewhella J, Guss JM, Matthews JM
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
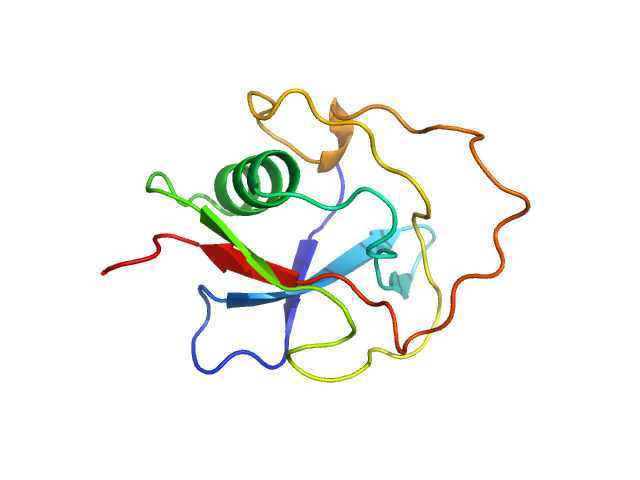
| Sample: |
Mouse Neurotrypsin Scavenger Receptor Cysteine-Rich Domain 3 monomer, 13 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 0.1 M NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 20
|
Structural characterization of the third scavenger receptor cysteine-rich domain of murine neurotrypsin.
Protein Sci 28(4):746-755 (2019)
Canciani A, Catucci G, Forneris F
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQP6_fit1_model1.png)
|
| Sample: |
Circularized Membrane scaffolding protein 1 E3 D1, 62 kDa protein
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) None, lipid
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 5
|
Circularized and solubility‐enhanced MSP
s facilitate simple and high‐yield production of stable nanodiscs for studies of membrane proteins in solution
The FEBS Journal 286(9):1734-1751 (2019)
Johansen N, Tidemand F, Nguyen T, Rand K, Pedersen M, Arleth L
|
| RgGuinier |
5.8 |
nm |
| Dmax |
14.5 |
nm |
|
|
|
|
|
|
|
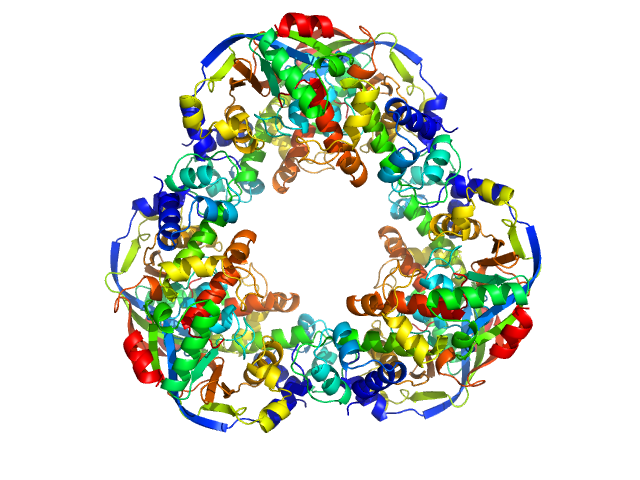
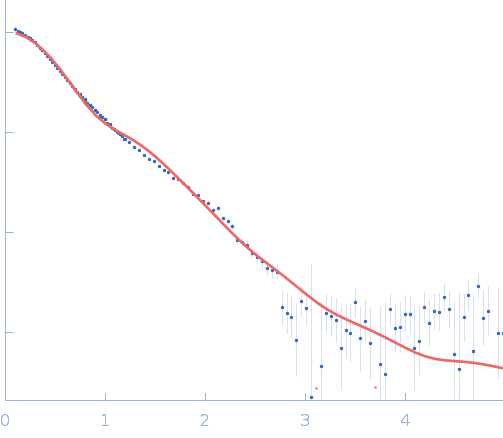
| Sample: |
Mycobacterial cidal toxin hexamer, 121 kDa Mycobacterium tuberculosis protein
Mycobacterial cidal antitoxin hexamer, 76 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
100 mM HEPES, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
An NAD+ Phosphorylase Toxin Triggers Mycobacterium tuberculosis Cell Death.
Mol Cell (2019)
Freire DM, Gutierrez C, Garza-Garcia A, Grabowska AD, Sala AJ, Ariyachaokun K, Panikova T, Beckham KSH, Colom A, Pogenberg V, Cianci M, Tuukkanen A, Boudehen YM, Peixoto A, Botella L, Svergun DI, Schnappinger D, Schneider TR, Genevaux P, de Carvalho LPS, Wilmanns M, Parret AHA, Neyrolles O
|
| RgGuinier |
4.1 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
262 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 11
|
NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation unique to this diflavin reductase
Journal of Structural Biology 205(2):170-179 (2019)
Tavolieri A, Murray D, Askenasy I, Pennington J, McGarry L, Stanley C, Stroupe M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
60 |
nm3 |
|
|