|
|
|
|
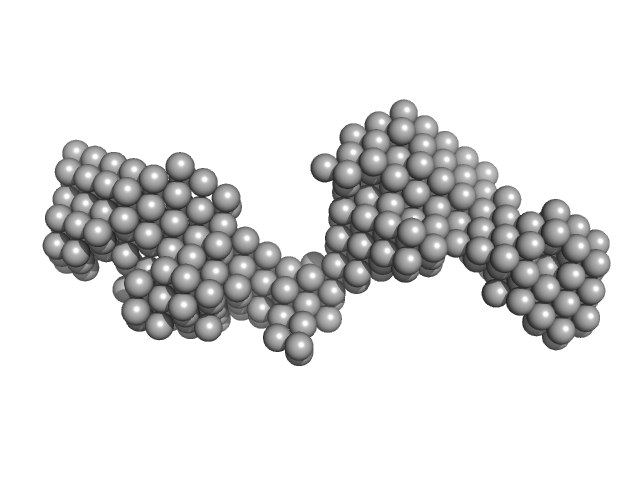
|
| Sample: |
Tegument protein UL37 monomer, 49 kDa Suid alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Inner tegument protein monomer, 62 kDa Human alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
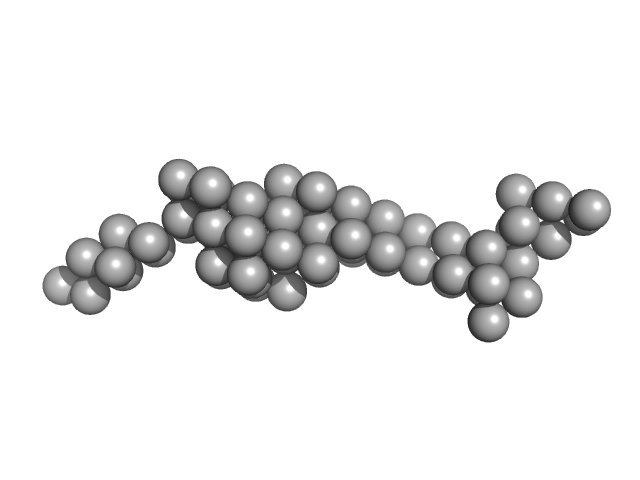
|
| Sample: |
Tegument protein UL37 monomer, 43 kDa Suid alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
|
|
|
|
|
|
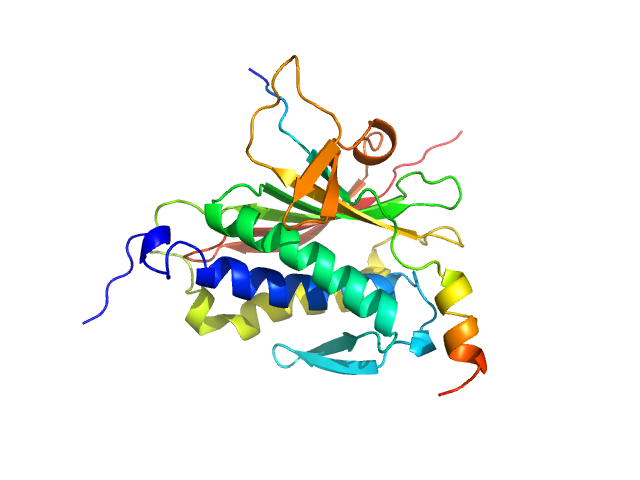
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B monomer, 24 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol, pH: 8.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
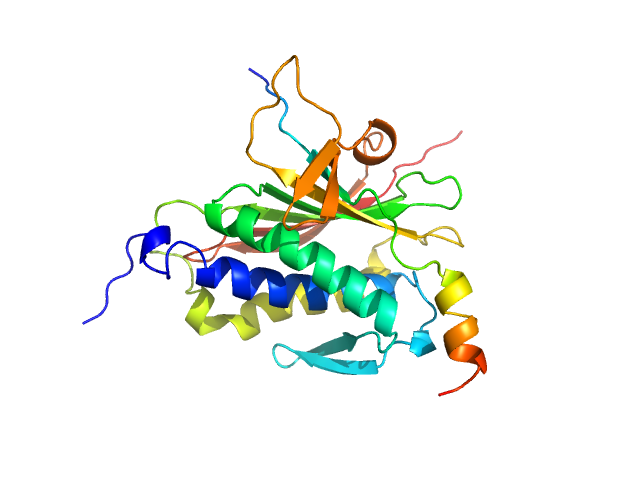
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B monomer, 24 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol, pH: 8.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
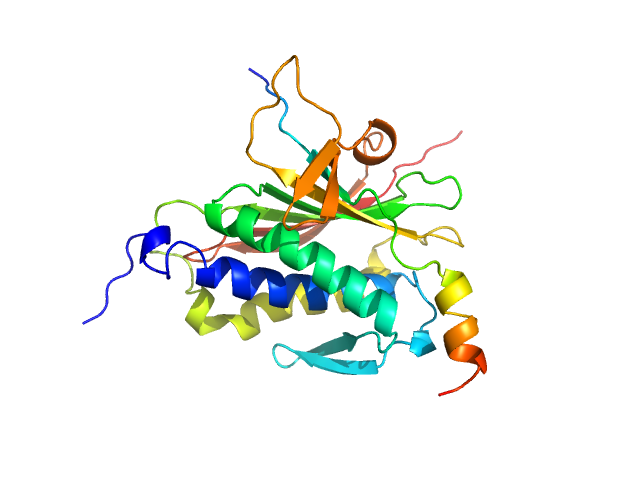
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B monomer, 24 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol, pH: 8.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
|
|
|
|
|
|
|
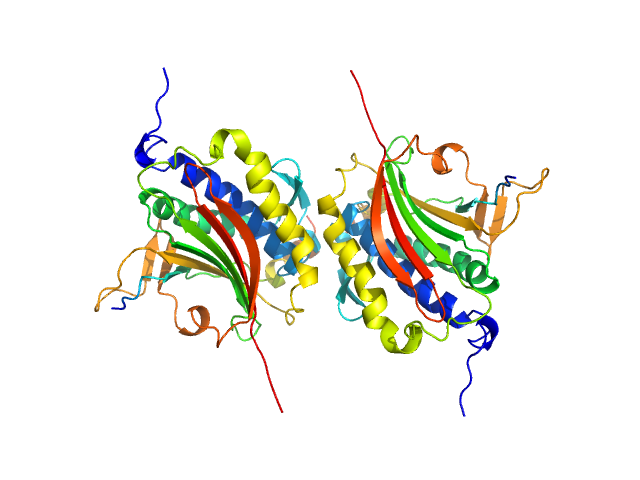
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
|
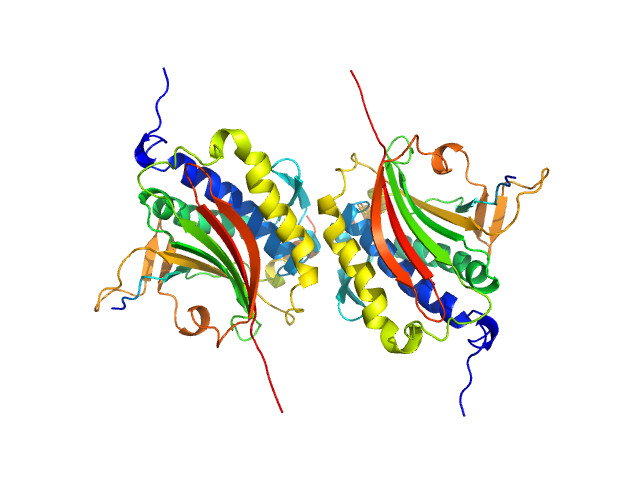
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitotic spindle assembly checkpoint protein MAD2B dimer, 49 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
DNA polymerase zeta catalytic subunit monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 10 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 14
|
Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex.
Proc Natl Acad Sci U S A 115(35):E8191-E8200 (2018)
Rizzo AA, Vassel FM, Chatterjee N, D'Souza S, Li Y, Hao B, Hemann MT, Walker GC, Korzhnev DM
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
105 |
nm3 |
|
|