|
|
|
|

|
| Sample: |
Human Filamin A Ig-like domains 16-17 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 50 mM NaCl 10 mM DTT, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Sep 30
|
Skeletal Dysplasia Mutations Effect on Human Filamins' Structure and Mechanosensing.
Sci Rep 7(1):4218 (2017)
Seppälä J, Bernardi RC, Haataja TJK, Hellman M, Pentikäinen OT, Schulten K, Permi P, Ylänne J, Pentikäinen U
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human Filamin A Ig-like domains 16-17* monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 50 mM NaCl 10 mM DTT, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Sep 30
|
Skeletal Dysplasia Mutations Effect on Human Filamins' Structure and Mechanosensing.
Sci Rep 7(1):4218 (2017)
Seppälä J, Bernardi RC, Haataja TJK, Hellman M, Pentikäinen OT, Schulten K, Permi P, Ylänne J, Pentikäinen U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
|
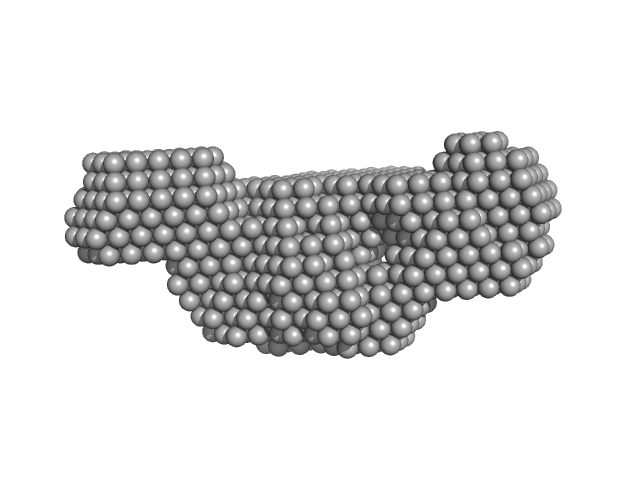
| Sample: |
Bcl-2-like protein FPV039 monomer, 17 kDa Fowlpox virus protein
Uncharacterized protein (BAK1) monomer, 3 kDa Gallus gallus protein
|
| Buffer: |
20 mM trisodium citrate pH, 200 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Oct 2
|
Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039.
J Biol Chem 292(22):9010-9021 (2017)
Anasir MI, Caria S, Skinner MA, Kvansakul M
|
|
|
|
|
|
|
|
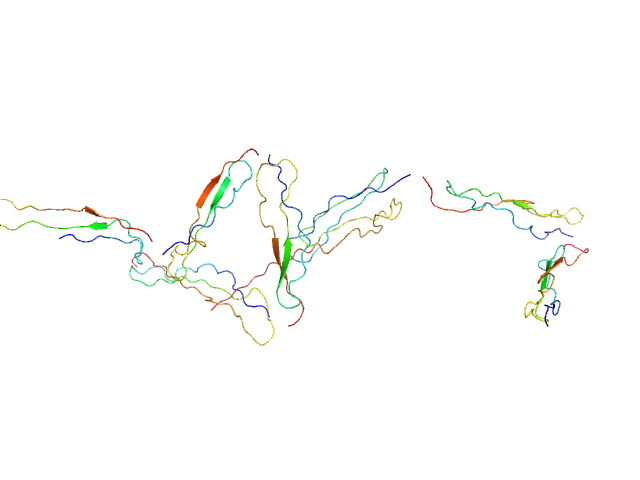
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Jul 2
|
TIA-1 RRM23 binding and recognition of target oligonucleotides.
Nucleic Acids Res 45(8):4944-4957 (2017)
Waris S, García-Mauriño SM, Sivakumaran A, Beckham SA, Loughlin FE, Gorospe M, Díaz-Moreno I, Wilce MCJ, Wilce JA
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
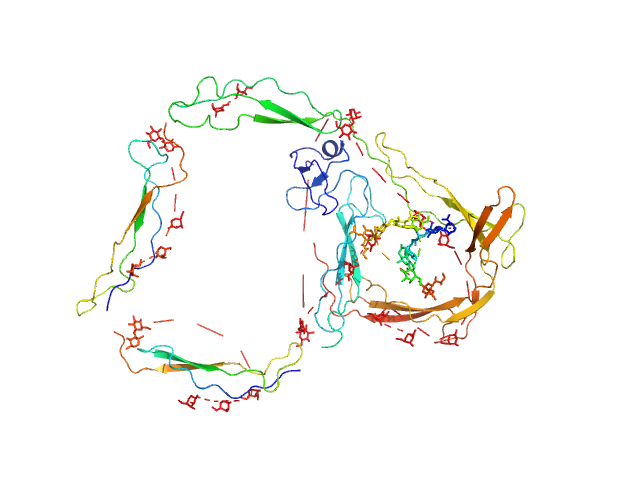
| Sample: |
Human recombinant Properdin TSR 0-3 monomer, 25 kDa Homo sapiens protein
Human recombinant Properdin TSR 4-6 monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Aug 16
|
Functional and structural insight into properdin control of complement alternative pathway amplification.
EMBO J 36(8):1084-1099 (2017)
Pedersen DV, Roumenina L, Jensen RK, Gadeberg TA, Marinozzi C, Picard C, Rybkine T, Thiel S, Sørensen UB, Stover C, Fremeaux-Bacchi V, Andersen GR
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
E244K Human Properdin monomer, 49 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 27
|
Functional and structural insight into properdin control of complement alternative pathway amplification.
EMBO J 36(8):1084-1099 (2017)
Pedersen DV, Roumenina L, Jensen RK, Gadeberg TA, Marinozzi C, Picard C, Rybkine T, Thiel S, Sørensen UB, Stover C, Fremeaux-Bacchi V, Andersen GR
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
ESX-5 type VII secretion system protein EccC5 monomer, 142 kDa Mycobacterium xenopi RIVM700367 protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 5%(v/v) Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 15
|
Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis.
Nat Microbiol 2:17047 (2017)
Beckham KS, Ciccarelli L, Bunduc CM, Mertens HD, Ummels R, Lugmayr W, Mayr J, Rettel M, Savitski MM, Svergun DI, Bitter W, Wilmanns M, Marlovits TC, Parret AH, Houben EN
|
| RgGuinier |
6.2 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
255 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes , 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.1 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 µM GppNHp, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes , 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Mar 12
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
70 |
nm3 |
|
|