|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 µM GppNHp, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes , 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Mar 12
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 µM GppNHp, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Mar 12
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Toxin CcdB dimer, 23 kDa Escherichia coli protein
Antitoxin CcdA dimer, 17 kDa Escherichia coli protein
Toxin CcdB dimer, 23 kDa Escherichia coli protein
|
| Buffer: |
10 mM Tris 50 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Jul 24
|
Molecular mechanism governing ratio-dependent transcription regulation in the ccdAB operon.
Nucleic Acids Res 45(6):2937-2950 (2017)
Vandervelde A, Drobnak I, Hadži S, Sterckx YG, Welte T, De Greve H, Charlier D, Efremov R, Loris R, Lah J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 uM GDP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
94673 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 uM GDP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, on 2015 Feb 1
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
70673 |
nm3 |
|
|
|
|
|
|

|
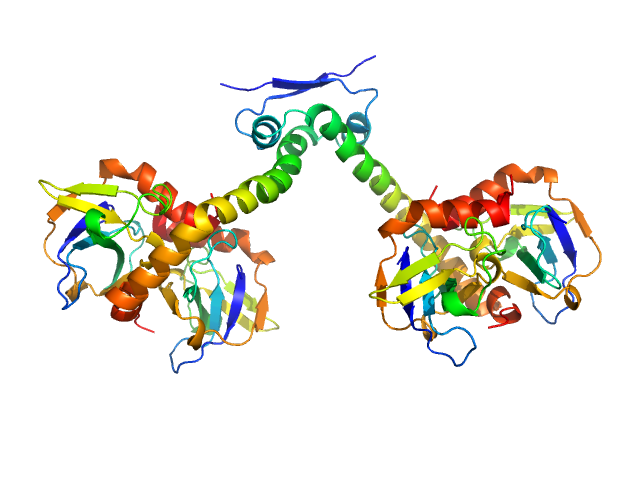
| Sample: |
Type-2 restriction enzyme AgeI monomer, 31 kDa Thalassobius gelatinovorus protein
|
| Buffer: |
10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM CaCl₂, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 14
|
Restriction endonuclease AgeI is a monomer which dimerizes to cleave DNA.
Nucleic Acids Res 45(6):3547-3558 (2017)
Tamulaitiene G, Jovaisaite V, Tamulaitis G, Songailiene I, Manakova E, Zaremba M, Grazulis S, Xu SY, Siksnys V
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
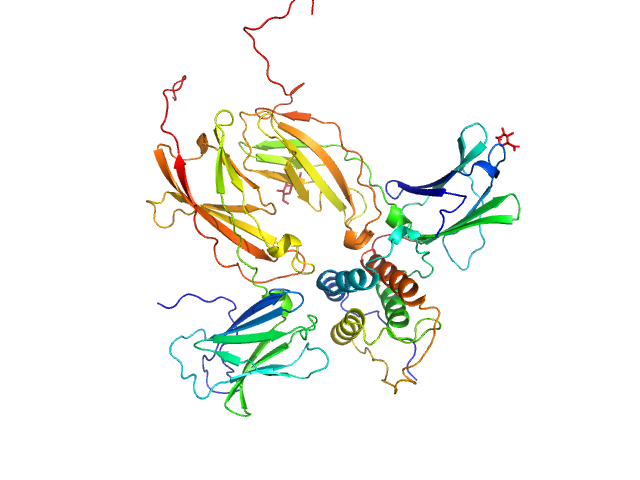
| Sample: |
Thymic stromal lymphopoietin monomer, 15 kDa Homo sapiens protein
Interleukin-7 receptor subunit alpha monomer, 26 kDa Homo sapiens protein
Cytokine receptor-like factor 2 monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 8
|
Structure and antagonism of the receptor complex mediated by human TSLP in allergy and asthma.
Nat Commun 8:14937 (2017)
Verstraete K, Peelman F, Braun H, Lopez J, Van Rompaey D, Dansercoer A, Vandenberghe I, Pauwels K, Tavernier J, Lambrecht BN, Hammad H, De Winter H, Beyaert R, Lippens G, Savvides SN
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
|
|
|
|
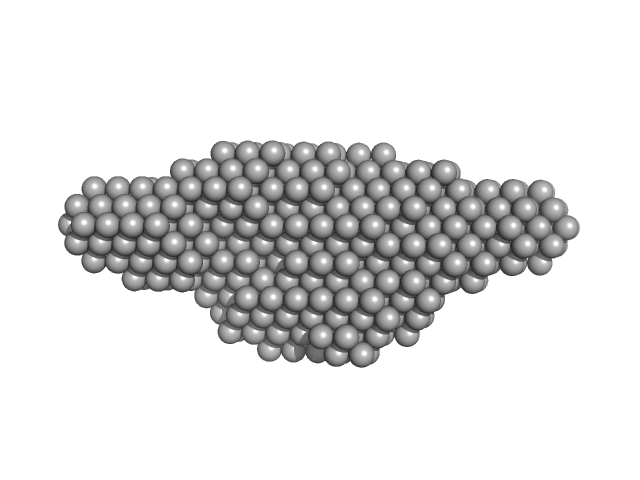
|
| Sample: |
Leishmania braziliensis heat shock protein 90 (Hsp90) dimer, 166 kDa Leishmania braziliensis protein
|
| Buffer: |
25 Tris mM 100 mM NaCl 1 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2011 Sep 1
|
Insights on the structural dynamics of Leishmania braziliensis Hsp90 molecular chaperone by small angle X-ray scattering.
Int J Biol Macromol 97:503-512 (2017)
Seraphim TV, Silva KP, Dores-Silva PR, Barbosa LR, Borges JC
|
| RgGuinier |
5.3 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
380 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Leishmania braziliensis heat shock protein 90 (Hsp90) N domain monomer, 26 kDa Leishmania braziliensis protein
|
| Buffer: |
25 Tris mM 100 mM NaCl 1 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2011 Sep 1
|
Insights on the structural dynamics of Leishmania braziliensis Hsp90 molecular chaperone by small angle X-ray scattering.
Int J Biol Macromol 97:503-512 (2017)
Seraphim TV, Silva KP, Dores-Silva PR, Barbosa LR, Borges JC
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
40 |
nm3 |
|
|