|
|
|
|

|
| Sample: |
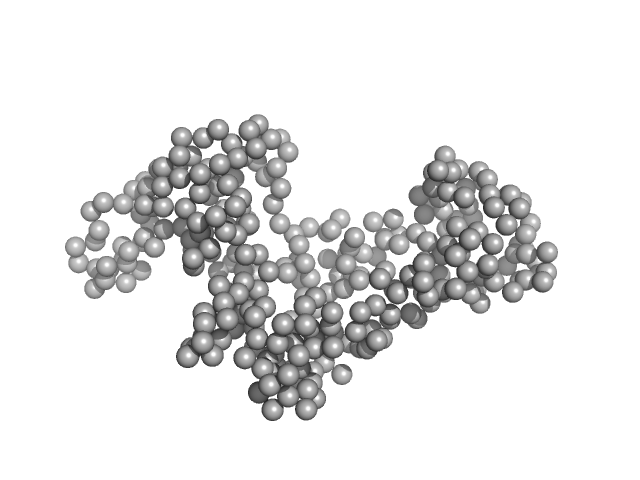
Leishmania braziliensis heat shock protein 90 (Hsp90) N and M domains monomer, 62 kDa Leishmania braziliensis protein
|
| Buffer: |
25 Tris mM 100 mM NaCl 1 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2011 Sep 1
|
Insights on the structural dynamics of Leishmania braziliensis Hsp90 molecular chaperone by small angle X-ray scattering.
Int J Biol Macromol 97:503-512 (2017)
Seraphim TV, Silva KP, Dores-Silva PR, Barbosa LR, Borges JC
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
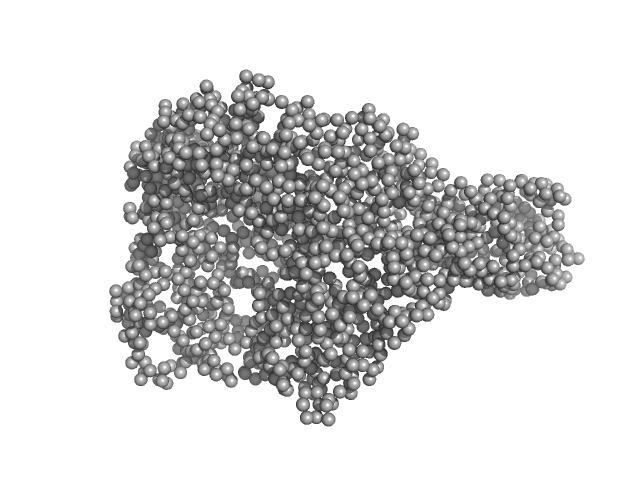
Chromodomain helicase DNA binding domain monomer, 31 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 20
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
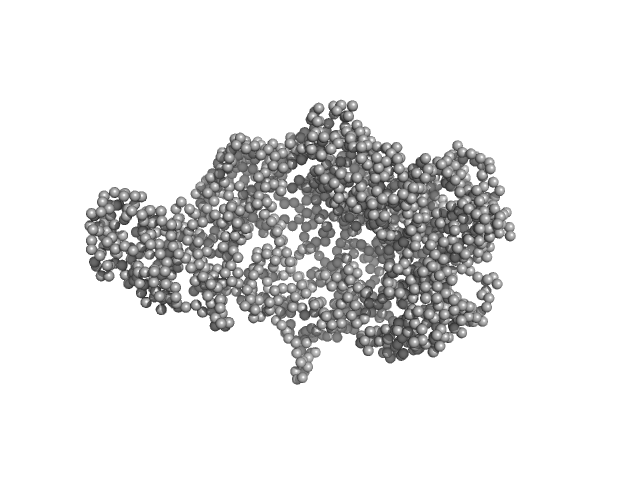
Chromodomain helicase DNA binding domain monomer, 135 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2008 Nov 30
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
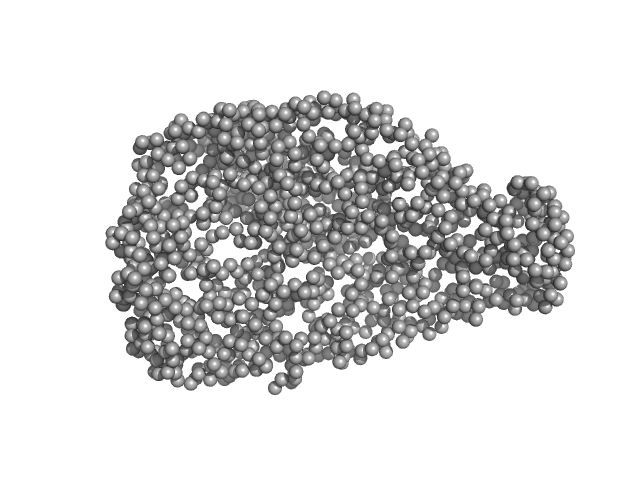
Chromodomain helicase DNA binding domain monomer, 150 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Nov 30
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
340 |
nm3 |
|
|
|
|
|
|
|
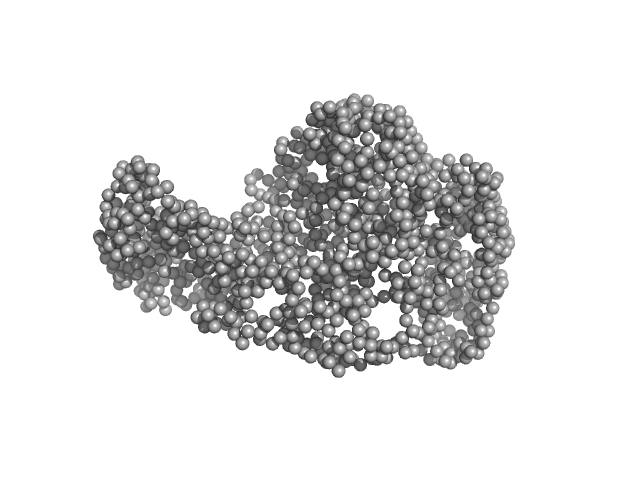
| Sample: |
Chromodomain helicase DNA binding domain monomer, 102 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Nov 30
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
4.1 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Chromodomain helicase DNA binding domain monomer, 117 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Nov 30
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
4.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
228 |
nm3 |
|
|
|
|
|
|
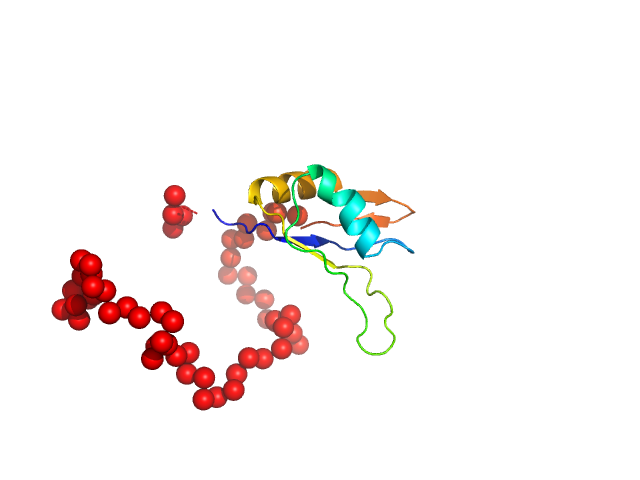
|
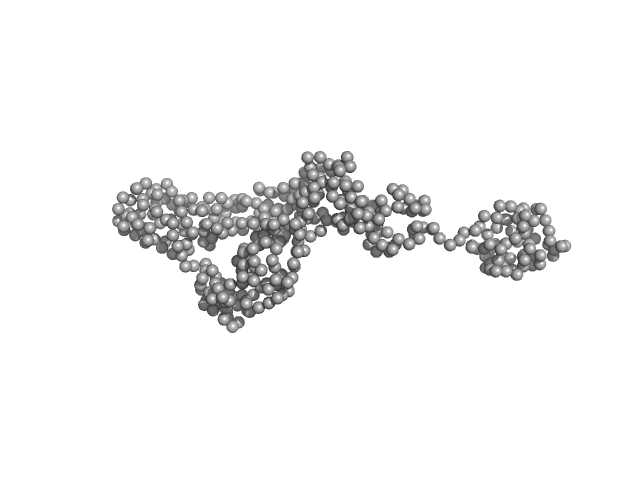
| Sample: |
Ribosome biogenesis protein 15 monomer, 17 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2013 Apr 25
|
Structural analysis reveals the flexible C-terminus of Nop15 undergoes rearrangement to recognize a pre-ribosomal RNA folding intermediate.
Nucleic Acids Res 45(5):2829-2837 (2017)
Zhang J, Gonzalez LE, Hall TMT
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endonuclease 8-like 1 monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
25mM HEPES 300mM NaCl 1mM DTT 10% Glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Mar 13
|
Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase.
Nucleic Acids Res 45(5):2897-2909 (2017)
Prakash A, Moharana K, Wallace SS, Doublié S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
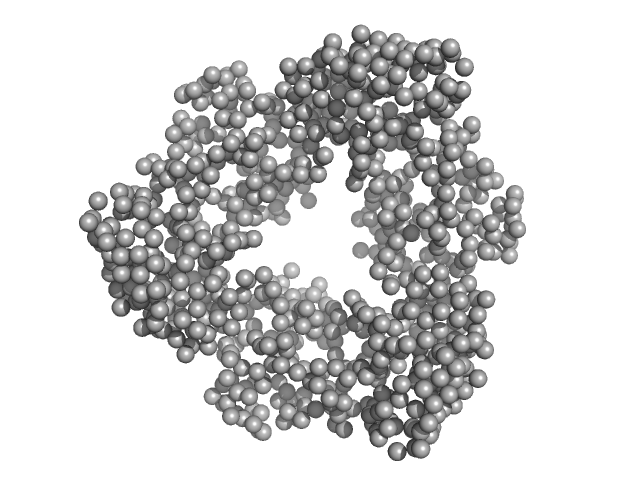
|
| Sample: |
Proliferating cell nuclear antigen trimer, 89 kDa Homo sapiens protein
|
| Buffer: |
25mM HEPES 100mM NaCl 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Jan 20
|
Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase.
Nucleic Acids Res 45(5):2897-2909 (2017)
Prakash A, Moharana K, Wallace SS, Doublié S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|
|
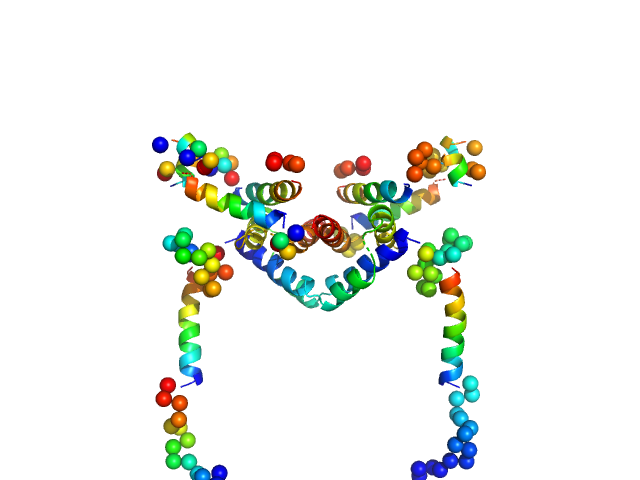
| Sample: |
Apoptosis regulator BAX (Bcl-2 associated X) dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
20mM sodium phosphate 100mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 May 28
|
Oligomerization process of Bcl-2 associated X protein revealed from intermediate structures in solution.
Phys Chem Chem Phys 19(11):7947-7954 (2017)
Shih O, Yeh YQ, Liao KF, Sung TC, Chiang YW, Jeng US
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
86 |
nm3 |
|
|