|
|
|
|
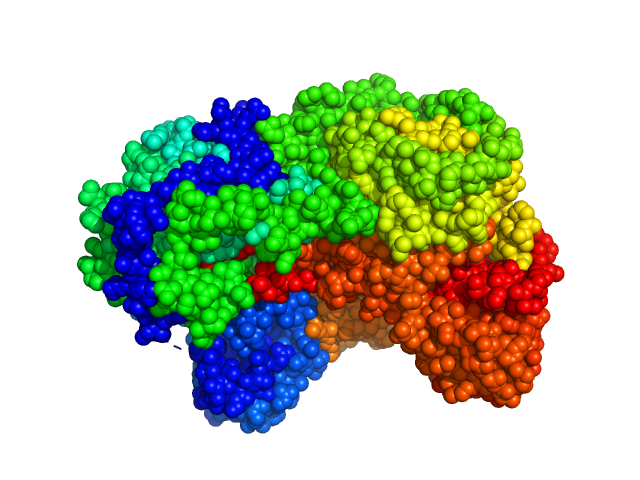
|
| Sample: |
Sinorhizobium meliloti (SmPutA) monomer, 132 kDa Sinorhizobium meliloti protein
|
| Buffer: |
50 mM Tris, 1% (v/v) glycerol, 0.5 mM THP, and 50 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Mar 27
|
Structures of Proline Utilization A (PutA) Reveal the Fold and Functions of the Aldehyde Dehydrogenase Superfamily Domain of Unknown Function.
J Biol Chem 291(46):24065-24075 (2016)
Luo M, Gamage TT, Arentson BW, Schlasner KN, Becker DF, Tanner JJ
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
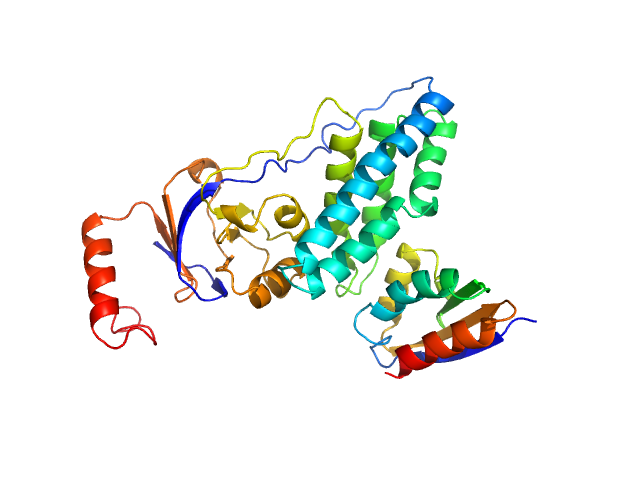
|
| Sample: |
Phosphocarrier protein NPr monomer, 9 kDa Escherichia coli protein
Phosphoenolpyruvate-protein phosphotransferase PtsP monomer, 28 kDa Escherichia coli protein
|
| Buffer: |
10 mM Tris 150 mM NaCl 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, NIDDK, NIH on 2015 Jan 30
|
Structure of the NPr:EINNtr Complex: Mechanism for Specificity in Paralogous Phosphotransferase Systems.
Structure 24(12):2127-2137 (2016)
Strickland M, Stanley AM, Wang G, Botos I, Schwieters CD, Buchanan SK, Peterkofsky A, Tjandra N
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
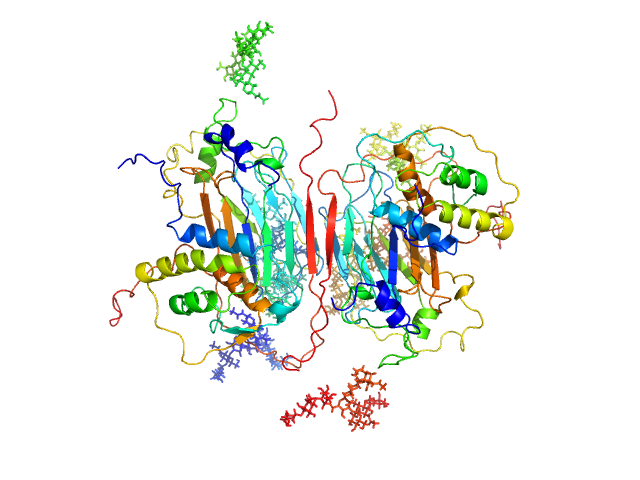
|
| Sample: |
GM-CSF/IL-2 inhibition factor tetramer, 120 kDa Orf virus protein
Granulocyte-macrophage colony-stimulating factor dimer, 29 kDa Ovis aries protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Sep 10
|
Structural basis of GM-CSF and IL-2 sequestration by the viral decoy receptor GIF.
Nat Commun 7:13228 (2016)
Felix J, Kandiah E, De Munck S, Bloch Y, van Zundert GC, Pauwels K, Dansercoer A, Novanska K, Read RJ, Bonvin AM, Vergauwen B, Verstraete K, Gutsche I, Savvides SN
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
231 |
nm3 |
|
|
|
|
|
|
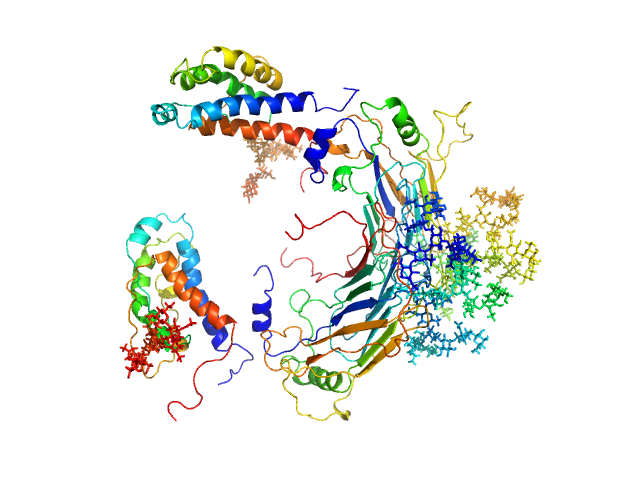
|
| Sample: |
GM-CSF/IL-2 inhibition factor tetramer, 120 kDa Orf virus protein
Interleukin-2 monomer, 16 kDa Ovis aries protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Sep 10
|
Structural basis of GM-CSF and IL-2 sequestration by the viral decoy receptor GIF.
Nat Commun 7:13228 (2016)
Felix J, Kandiah E, De Munck S, Bloch Y, van Zundert GC, Pauwels K, Dansercoer A, Novanska K, Read RJ, Bonvin AM, Vergauwen B, Verstraete K, Gutsche I, Savvides SN
|
| RgGuinier |
4.1 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
|
|
|
|
|
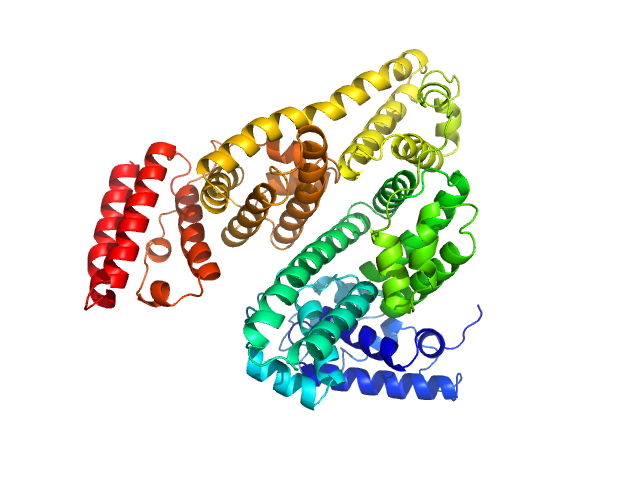
| Sample: |
Bovine serum albumin, monomer monomer, 66 kDa Bos taurus protein
|
| Buffer: |
25 mM Tris 150 mM NaCl 3% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 23
|
Preparing monodisperse macromolecular samples for successful biological small-angle X-ray and neutron-scattering experiments.
Nat Protoc 11(11):2122-2153 (2016)
Jeffries CM, Graewert MA, Blanchet CE, Langley DB, Whitten AE, Svergun DI
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
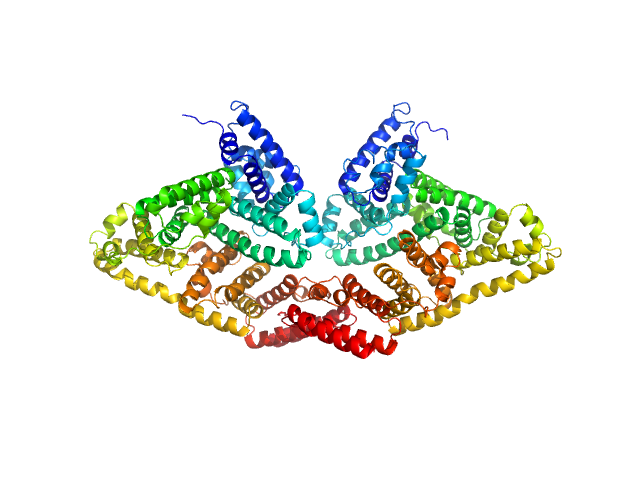
| Sample: |
Bovine serum albumin, dimer dimer, 133 kDa Bos taurus protein
|
| Buffer: |
25 mM Tris 150 mM NaCl 3% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 23
|
Preparing monodisperse macromolecular samples for successful biological small-angle X-ray and neutron-scattering experiments.
Nat Protoc 11(11):2122-2153 (2016)
Jeffries CM, Graewert MA, Blanchet CE, Langley DB, Whitten AE, Svergun DI
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
202 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cardiac myosin binding protein C: tri-helix bundle-C2 monomer, 15 kDa human sequence obtained … protein
|
| Buffer: |
150 mM NaCl, 10 mM MES, 2 mM TCEP, 1 mM NaN3 at 4°C, pH: 6.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 18
|
A Highly Conserved Yet Flexible Linker Is Part of a Polymorphic Protein-Binding Domain in Myosin-Binding Protein C.
Structure 24(11):2000-2007 (2016)
Michie KA, Kwan AH, Tung CS, Guss JM, Trewhella J
|
| RgGuinier |
1.9 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
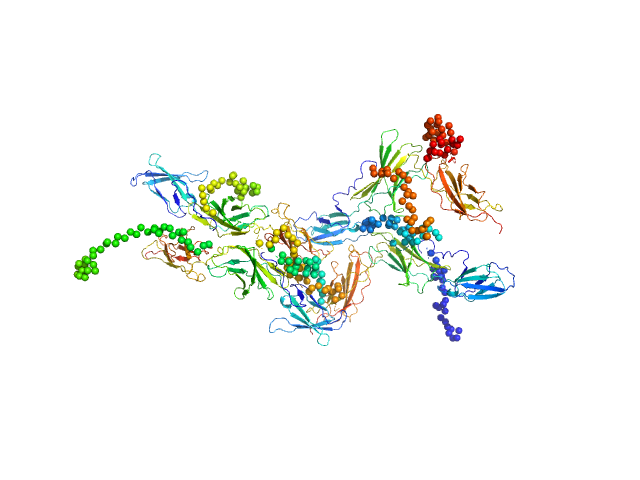
| Sample: |
Interleukin-6 receptor subunit alpha tetramer, 164 kDa Homo sapiens protein
Interleukin-6 receptor subunit alpha dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2014 Oct 1
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
4.9 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
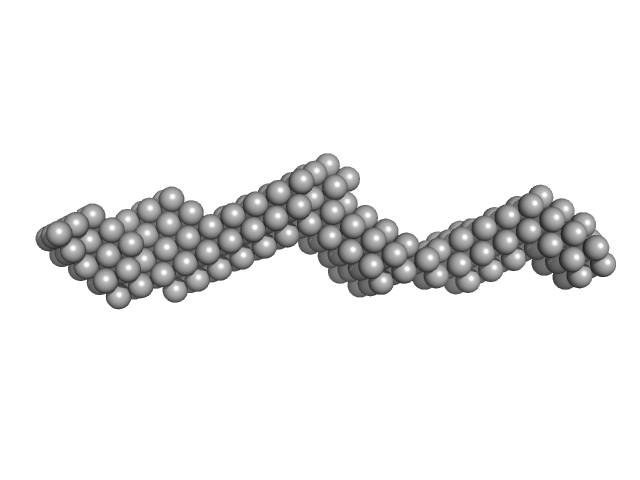
| Sample: |
Plakin domain fragment of Human plectin encompassing spectrin repeats SR3-SR4-SR5-SR6 and SH3 monomer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 26
|
The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape.
J Biol Chem 291(36):18643-62 (2016)
Ortega E, Manso JA, Buey RM, Carballido AM, Carabias A, Sonnenberg A, de Pereda JM
|
| RgGuinier |
5.1 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
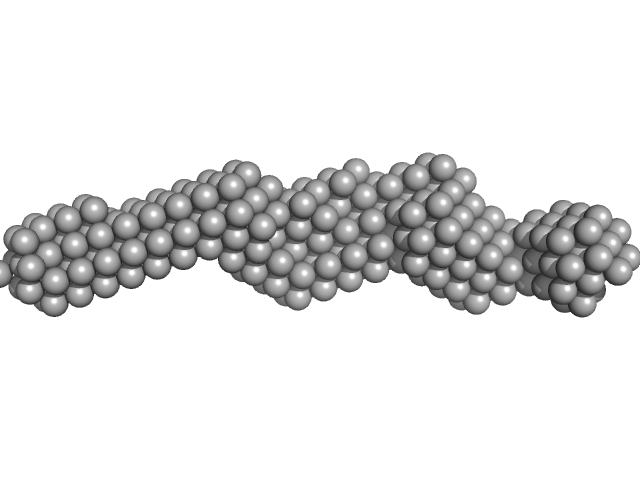
| Sample: |
Plakin domain fragment of Human plectin encompassing spectrin repeats SR7-SR8-SR9 monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 26
|
The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape.
J Biol Chem 291(36):18643-62 (2016)
Ortega E, Manso JA, Buey RM, Carballido AM, Carabias A, Sonnenberg A, de Pereda JM
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
57 |
nm3 |
|
|