|
|
|
|

|
| Sample: |
Human Chromatin Remodeler CHD4 (494-1353) monomer, 101 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Nov 13
|
The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4.
J Mol Biol 422(1):3-17 (2012)
Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, Du X, Bartke T, Zhang W, Edlich C, Berridge G, Chen Y, Burgess-Brown NA, Kouzarides T, Wiechens N, Owen-Hughes T, Svergun DI, Gileadi O, Laue ED
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Human Chromatin Remodeler CHD4 (685-1233) monomer, 63 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Nov 13
|
The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4.
J Mol Biol 422(1):3-17 (2012)
Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, Du X, Bartke T, Zhang W, Edlich C, Berridge G, Chen Y, Burgess-Brown NA, Kouzarides T, Wiechens N, Owen-Hughes T, Svergun DI, Gileadi O, Laue ED
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Human Chromatin Remodeler CHD4 (363-682) monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Nov 13
|
The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4.
J Mol Biol 422(1):3-17 (2012)
Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, Du X, Bartke T, Zhang W, Edlich C, Berridge G, Chen Y, Burgess-Brown NA, Kouzarides T, Wiechens N, Owen-Hughes T, Svergun DI, Gileadi O, Laue ED
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
|
|
|
|
|
|
|
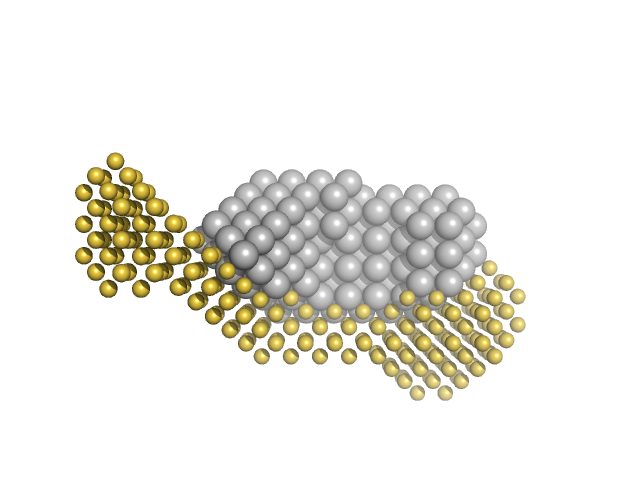
| Sample: |
Calmodulin-1 monomer, 17 kDa Xenopus laevis protein
Calcium ions tetramer, 0 kDa
Gag-Pol polyprotein monomer, 15 kDa Human immunodeficiency virus … protein
|
| Buffer: |
50 mM MOPS, 5 mM CaCl2, 2 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, Australian Nuclear Science and Technology Organisation on 2010 Jun 23
|
Calmodulin binds a highly extended HIV-1 MA protein that refolds upon its release.
Biophys J 103(3):541-549 (2012)
Taylor JE, Chow JYH, Jeffries CM, Kwan AH, Duff AP, Hamilton WA, Trewhella J
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
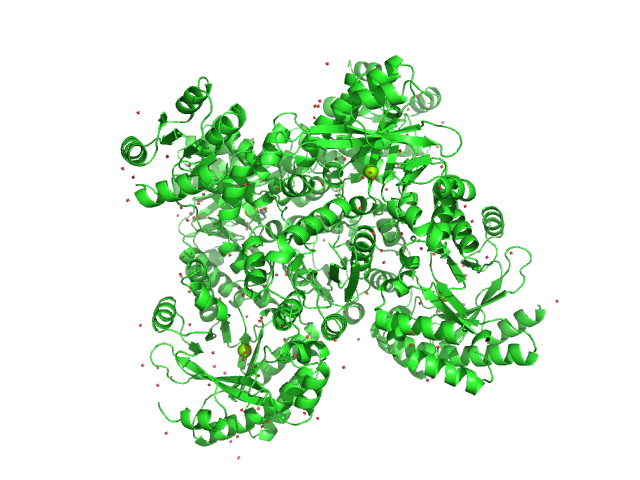
|
| Sample: |
RocR tetramer, 171 kDa Pseudomonas aeruginosa (strain … protein
|
| Buffer: |
50 mM Tris–HCl, 250 mM NaCl, 10 mM imidazole, 5% glycerol, 0.5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 9
|
Structural Insights into the Regulatory Mechanism of the Response Regulator RocR from Pseudomonas aeruginosa in Cyclic Di-GMP Signaling
Journal of Bacteriology 194(18):4837-4846 (2012)
Chen M, Kotaka M, Vonrhein C, Bricogne G, Rao F, Chuah M, Svergun D, Schneider G, Liang Z, Lescar J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
CYNEX4 FRET probe, (eYFP-AnnexinA4-eCFP) monomer, 93 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1 mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Sep 11
|
Conformational analysis of a genetically encoded FRET biosensor by SAXS.
Biophys J 102(12):2866-75 (2012)
Mertens HD, Piljić A, Schultz C, Svergun DI
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
135 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
CYNEX4 FRET probe, (eYFP-AnnexinA4-eCFP) T266D mutant monomer, 93 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Sep 11
|
Conformational analysis of a genetically encoded FRET biosensor by SAXS.
Biophys J 102(12):2866-75 (2012)
Mertens HD, Piljić A, Schultz C, Svergun DI
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
146 |
nm3 |
|
|
|
|
|
|
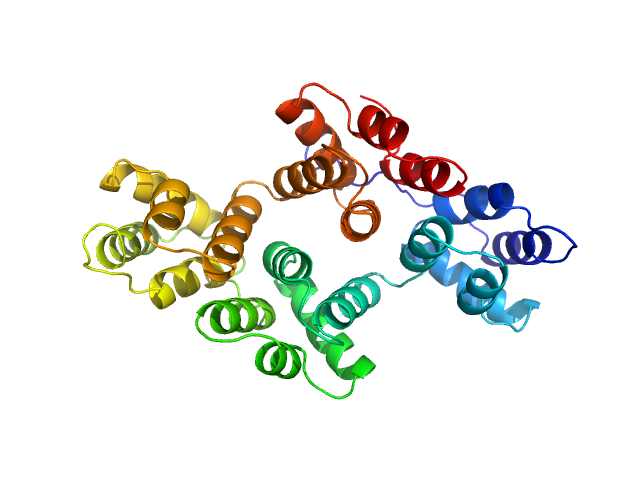
|
| Sample: |
Annexin-A4 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Sep 11
|
Conformational analysis of a genetically encoded FRET biosensor by SAXS.
Biophys J 102(12):2866-75 (2012)
Mertens HD, Piljić A, Schultz C, Svergun DI
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
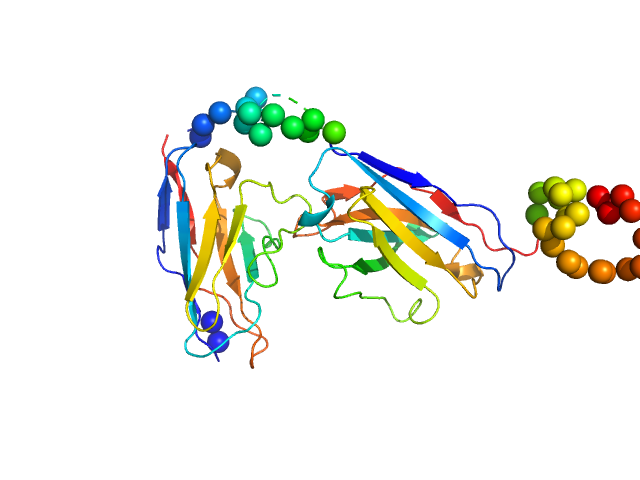
|
| Sample: |
Recombinant monoclonal anti-proNGF antibody in single chain Fv fragment (scFv) monomer, 29 kDa protein
|
| Buffer: |
phosphate buffered saline, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Oct 21
|
Direct intracellular selection and biochemical characterization of a recombinant anti-proNGF single chain antibody fragment.
Arch Biochem Biophys 522(1):26-36 (2012)
Paoletti F, Malerba F, Konarev PV, Visintin M, Scardigli R, Fasulo L, Lamba D, Svergun DI, Cattaneo A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Frataxin homolog, mitochondrial monomer, 14 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM HEPES, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Nov 28
|
The role of hydration in protein stability: comparison of the cold and heat unfolded states of Yfh1.
J Mol Biol 417(5):413-24 (2012)
Adrover M, Martorell G, Martin SR, Urosev D, Konarev PV, Svergun DI, Daura X, Temussi P, Pastore A
|
|
|