|
|
|
|
|
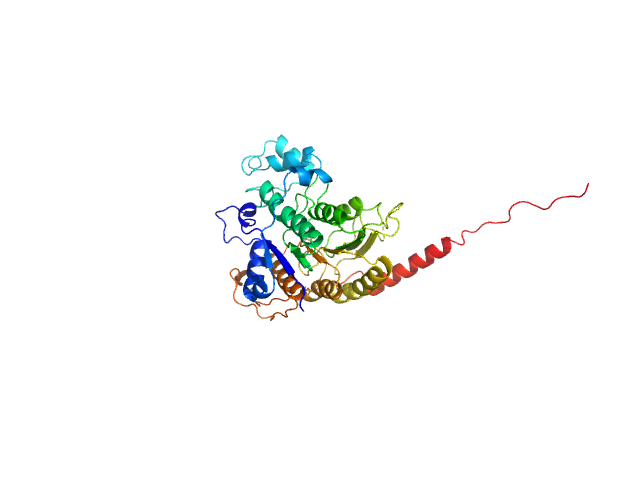
| Sample: |
Endoplasmin dimer, 169 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Jul 11
|
Visualization of conformational transition of GRP94 in solution.
Life Sci Alliance 7(2) (2024)
Sun S, Zhu R, Zhu M, Wang Q, Li N, Yang B
|
| RgGuinier |
5.7 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoplasmin dimer, 169 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Jul 11
|
Visualization of conformational transition of GRP94 in solution.
Life Sci Alliance 7(2) (2024)
Sun S, Zhu R, Zhu M, Wang Q, Li N, Yang B
|
| RgGuinier |
5.5 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histone deacetylase 7 monomer, 44 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 24
|
Structure-function analyses reveal Arabidopsis thaliana HDA7 to be an inactive histone deacetylase
Current Research in Structural Biology :100136 (2024)
Saharan K, Baral S, Shaikh N, Vasudevan D
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein DPCD monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2018 Nov 8
|
DPCD is a regulator of R2TP in ciliogenesis initiation through Akt signaling.
Cell Rep 43(2):113713 (2024)
Mao YQ, Seraphim TV, Wan Y, Wu R, Coyaud E, Bin Munim M, Mollica A, Laurent E, Babu M, Mennella V, Raught B, Houry WA
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Kinesin heavy chain (A515T) dimer, 221 kDa Drosophila melanogaster protein
|
| Buffer: |
25 mM HEPES/KOH, 150 mM NaCl, 1 mM MgCl2, 2 mM DTT, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Feb 19
|
Tropomyosin 1-I/C coordinates kinesin-1 and dynein motors during oskar mRNA transport.
Nat Struct Mol Biol (2024)
Heber S, McClintock MA, Simon B, Mehtab E, Lapouge K, Hennig J, Bullock SL, Ephrussi A
|
| RgGuinier |
10.0 |
nm |
| Dmax |
45.0 |
nm |
| VolumePorod |
920 |
nm3 |
|
|
|
|
|
|
|
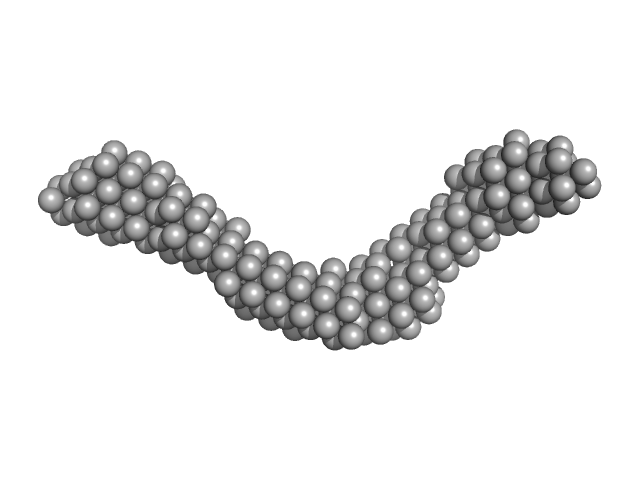
| Sample: |
Kinesin heavy chain (A515T) dimer, 221 kDa Drosophila melanogaster protein
GH09289p dimer, 96 kDa Drosophila melanogaster protein
|
| Buffer: |
25 mM HEPES/KOH, 150 mM NaCl, 1 mM MgCl2, 2 mM DTT, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Feb 19
|
Tropomyosin 1-I/C coordinates kinesin-1 and dynein motors during oskar mRNA transport.
Nat Struct Mol Biol (2024)
Heber S, McClintock MA, Simon B, Mehtab E, Lapouge K, Hennig J, Bullock SL, Ephrussi A
|
| RgGuinier |
8.7 |
nm |
| Dmax |
45.0 |
nm |
| VolumePorod |
840 |
nm3 |
|
|
|
|
|
|
|
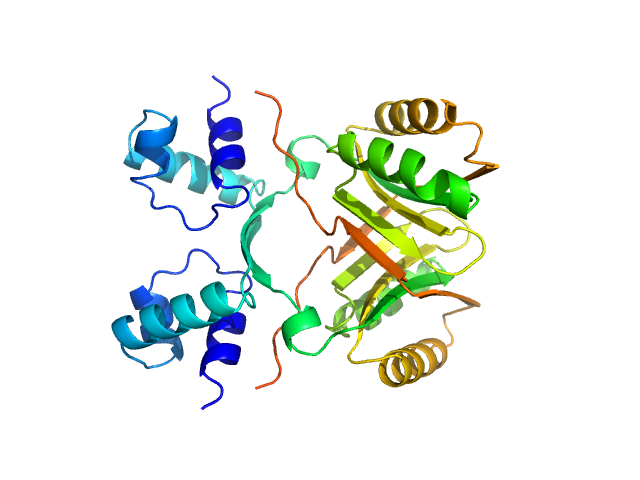
| Sample: |
Cadherin EGF LAG seven-pass G-type receptor 1 monomer, 49 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM KCl, 50 mM NaCl, 2 mM CaCl2, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Jul 1
|
CELSR1, a core planar cell polarity protein, features a weakly adhesive and flexible cadherin ectodomain.
Structure (2024)
Tamilselvan E, Sotomayor M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
AsnC family transcriptional regulator dimer, 36 kDa Haloferax mediterranei (strain … protein
|
| Buffer: |
50 mM Tris, 1.5 M NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 12
|
Global Lrp regulator protein from Haloferax mediterranei: Transcriptional analysis and structural characterization.
Int J Biol Macromol :129541 (2024)
Matarredona L, García-Bonete MJ, Guío J, Camacho M, Fillat MF, Esclapez J, Bonete MJ
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
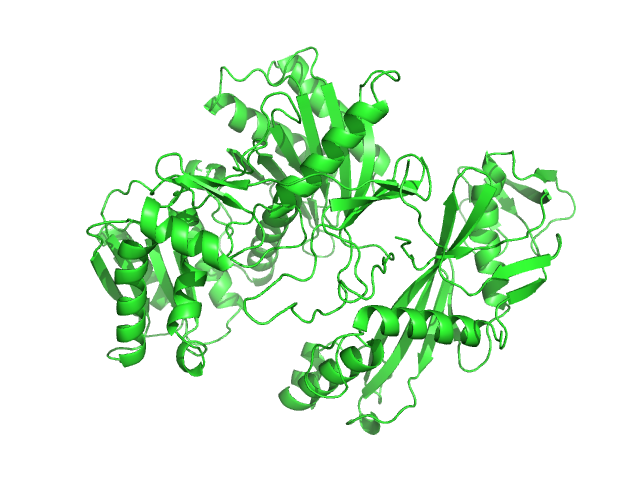
| Sample: |
Phosphocholinase AnkX monomer, 107 kDa Legionella pneumophila protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Oct 20
|
Legionella pneumophila effector AnkX
Wenhua Zhang
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Single-chain full Archaeoglobus fulgidus Argonaute variant monomer, 78 kDa Archaeoglobus fulgidus protein
|
| Buffer: |
20 mM TrisHCl pH 7.5, 500 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Aug 31
|
The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res (2024)
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
142 |
nm3 |
|
|