|
|
|
|
|
| Sample: |
Ribose import binding protein RbsB monomer, 30 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 10% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 15
|
SAXS/WAXS data of conformationally flexible ribose binding protein
Data in Brief :109932 (2023)
Choudhury J, Yonezawa K, Anu A, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ribose import binding protein RbsB monomer, 30 kDa Escherichia coli (strain … protein
Ribose monomer, 0 kDa
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 10% glycerol, 1 mM ribose, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 15
|
SAXS/WAXS data of conformationally flexible ribose binding protein
Data in Brief :109932 (2023)
Choudhury J, Yonezawa K, Anu A, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ribose import binding protein RbsB monomer, 30 kDa Escherichia coli (strain … protein
Ribose monomer, 0 kDa
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 10% glycerol, 1 mM ribose, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 15
|
SAXS/WAXS data of conformationally flexible ribose binding protein
Data in Brief :109932 (2023)
Choudhury J, Yonezawa K, Anu A, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ribose import binding protein RbsB monomer, 30 kDa Escherichia coli (strain … protein
Ribose monomer, 0 kDa
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 10% glycerol, 1 mM ribose, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 15
|
SAXS/WAXS data of conformationally flexible ribose binding protein
Data in Brief :109932 (2023)
Choudhury J, Yonezawa K, Anu A, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ribose import binding protein RbsB monomer, 30 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 10% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 9
|
SAXS/WAXS data of conformationally flexible ribose binding protein
Data in Brief :109932 (2023)
Choudhury J, Yonezawa K, Anu A, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ribose import binding protein RbsB monomer, 30 kDa Escherichia coli (strain … protein
Ribose monomer, 0 kDa
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 10% glycerol, 1 mM ribose, pH: 7 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 9
|
SAXS/WAXS data of conformationally flexible ribose binding protein
Data in Brief :109932 (2023)
Choudhury J, Yonezawa K, Anu A, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ribose import binding protein RbsB monomer, 30 kDa Escherichia coli (strain … protein
Ribose monomer, 0 kDa
|
| Buffer: |
Tris 50 mM, 50 mM NaCl, 10% glycerol, 5 mM ribose, pH: 7 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 9
|
SAXS/WAXS data of conformationally flexible ribose binding protein
Data in Brief :109932 (2023)
Choudhury J, Yonezawa K, Anu A, Shimizu N, Chaudhuri B
|
| RgGuinier |
1.9 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
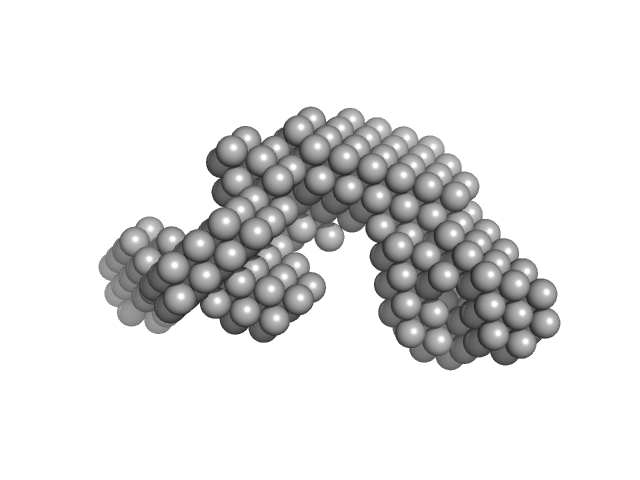
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
296 |
nm3 |
|
|
|
|
|
|
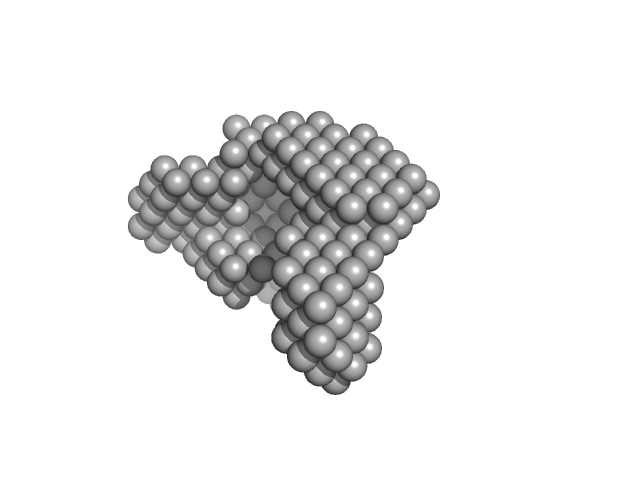
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 336-418 monomer, 9 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
279 |
nm3 |
|
|
|
|
|
|
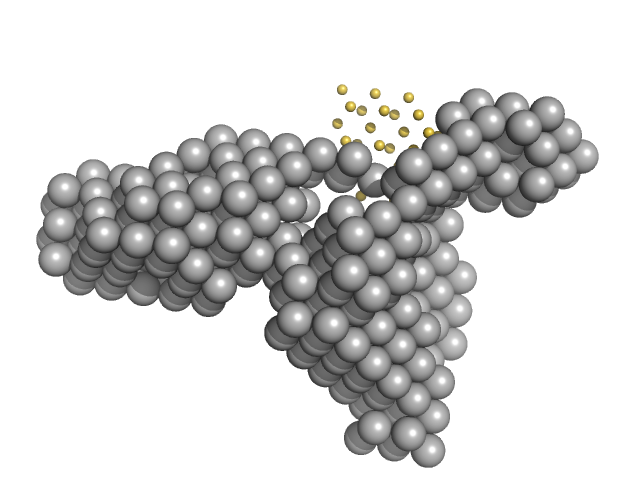
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 225-418 monomer, 21 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
309 |
nm3 |
|
|