|
|
|
|
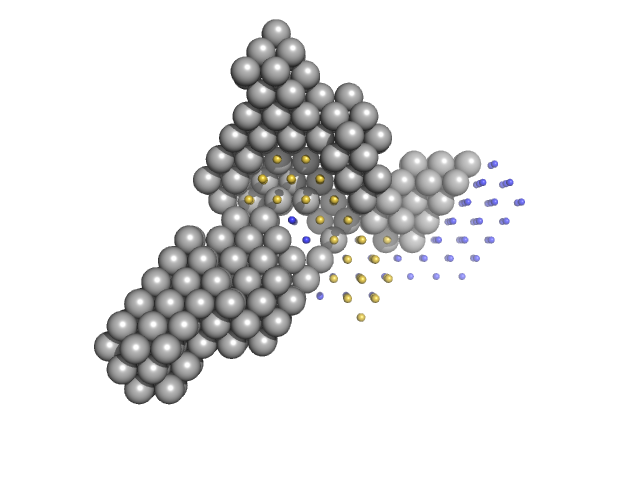
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 225-418 monomer, 21 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 776-898 monomer, 14 kDa Chaetomium thermophilum protein
SMC hinge domain-containing protein, 263-466 monomer, 22 kDa Chaetomium thermophilum protein
SMC hinge domain-containing protein, 1367-1542 monomer, 20 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
355 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ubiquilin-1 dimer, 125 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Mar 22
|
Short disordered N-termini & proline-rich domain are major regulators of UBQLN1/2/4 phase separation.
Biophys J (2023)
Dao TP, Rajendran A, Galagedera SKK, Haws W, Castañeda CA
|
| RgGuinier |
4.6 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
264 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ubiquilin-2 dimer, 131 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 8
|
Short disordered N-termini & proline-rich domain are major regulators of UBQLN1/2/4 phase separation.
Biophys J (2023)
Dao TP, Rajendran A, Galagedera SKK, Haws W, Castañeda CA
|
| RgGuinier |
5.2 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ubiquilin-4 dimer, 128 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 26
|
Short disordered N-termini & proline-rich domain are major regulators of UBQLN1/2/4 phase separation.
Biophys J (2023)
Dao TP, Rajendran A, Galagedera SKK, Haws W, Castañeda CA
|
| RgGuinier |
5.1 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
333 |
nm3 |
|
|
|
|
|
|
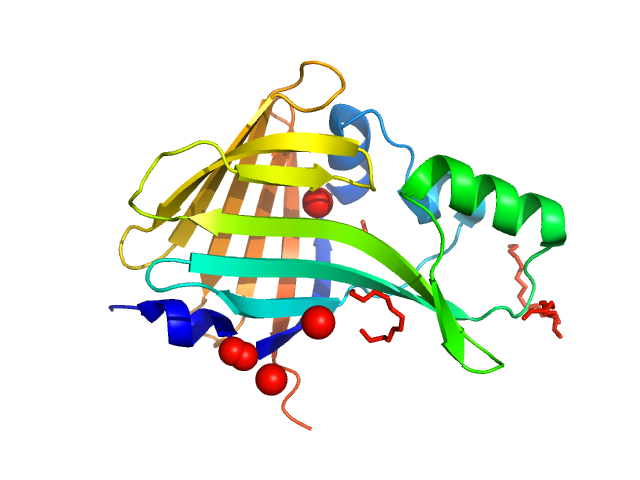
|
| Sample: |
Oplophorus-luciferin 2-monooxygenase catalytic subunit monomer, 20 kDa Oplophorus gracilirostris protein
|
| Buffer: |
10 mM Tris-HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2021 May 27
|
Illuminating the mechanism and allosteric behavior of NanoLuc luciferase.
Nat Commun 14(1):7864 (2023)
Nemergut M, Pluskal D, Horackova J, Sustrova T, Tulis J, Barta T, Baatallah R, Gagnot G, Novakova V, Majerova M, Sedlackova K, Marques SM, Toul M, Damborsky J, Prokop Z, Bednar D, Janin YL, Marek M
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase DTX3L tetramer, 46 kDa Homo sapiens protein
|
| Buffer: |
30 mM HEPES, 350 mM NaCl, 5% (v/v) glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Dec 6
|
Oligomerisation mediated by the D2 domain of DTX3L is critical for DTX3L-PARP9 reading function of mono-ADP-ribosylated androgen receptor.
bioRxiv (2023)
Vela-Rodríguez C, Yang C, Alanen HI, Eki R, Abbas TA, Maksimainen MM, Glumoff T, Duman R, Wagner A, Paschal BM, Lehtiö L
|
| RgGuinier |
2.9 |
nm |
| Dmax |
112.0 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
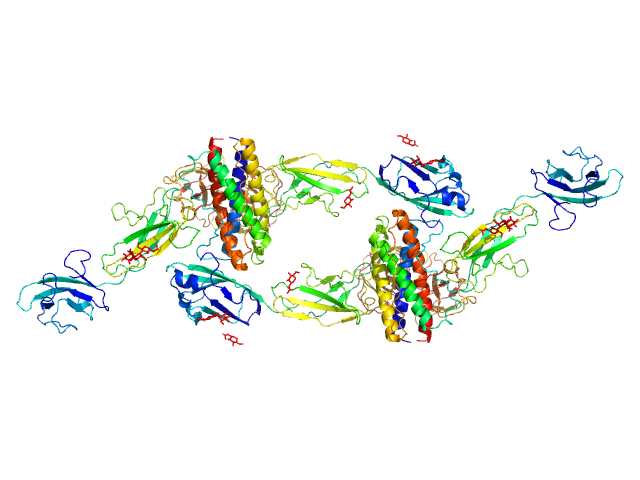
|
| Sample: |
Interleukin-6 receptor subunit beta dimer, 69 kDa Homo sapiens protein
Interleukin-11 dimer, 36 kDa Homo sapiens protein
Interleukin-11 receptor subunit alpha dimer, 64 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
5.3 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
405 |
nm3 |
|
|
|
|
|
|
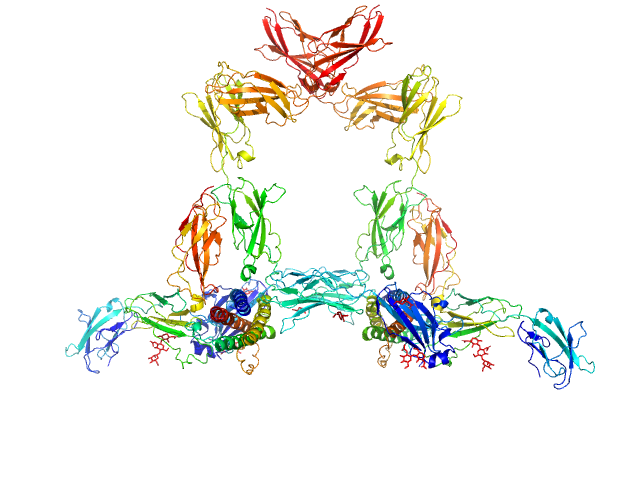
|
| Sample: |
Interleukin-11 dimer, 36 kDa Homo sapiens protein
Interleukin-11 receptor subunit alpha dimer, 64 kDa Homo sapiens protein
Interleukin-6 receptor subunit beta dimer, 134 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 28
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
6.2 |
nm |
| Dmax |
20.7 |
nm |
| VolumePorod |
710 |
nm3 |
|
|
|
|
|
|
|
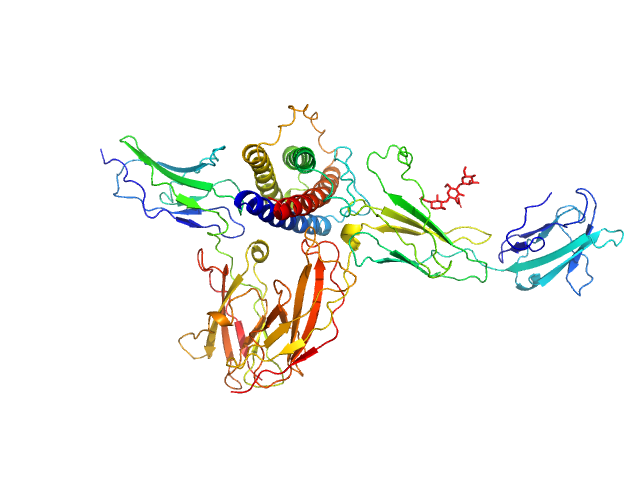
| Sample: |
Interleukin-11 receptor subunit alpha monomer, 32 kDa Homo sapiens protein
Interleukin 11 monomer, 18 kDa Homo sapiens protein
Interleukin-6 receptor subunit beta monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|
|
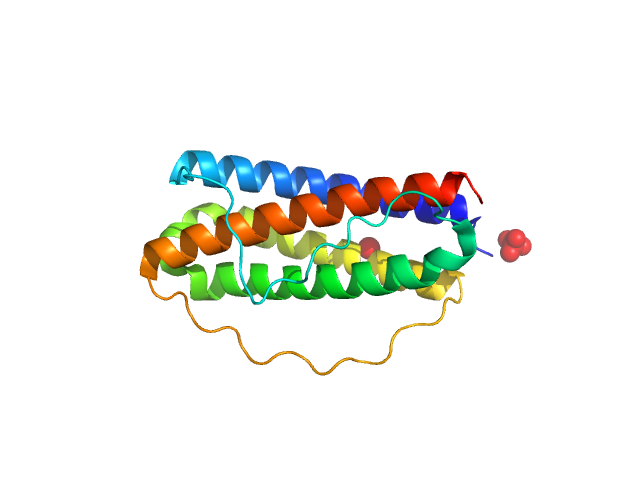
| Sample: |
Interleukin-11 (W168A) monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 28
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
22 |
nm3 |
|
|