|
|
|
|
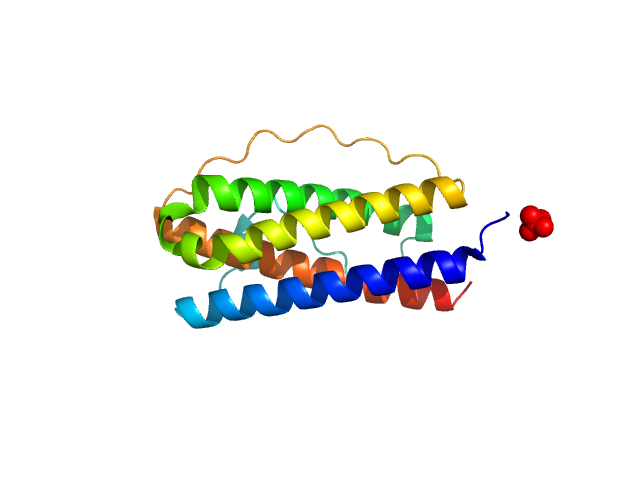
|
| Sample: |
Interleukin 11 Mutein monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Interleukin-11 receptor subunit alpha monomer, 32 kDa Homo sapiens protein
Interleukin 11 Mutein monomer, 18 kDa Homo sapiens protein
Interleukin-6 receptor subunit beta monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
4.4 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Queuine tRNA-ribosyltransferase catalytic subunit 1 monomer, 44 kDa Homo sapiens protein
Queuine tRNA-ribosyltransferase accessory subunit 2 monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 3% (w/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Dec 1
|
Structural and functional investigation of tRNA guanine transglycosylase
University of Göttingen Dissertation - (2023)
Katharina Sievers
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|
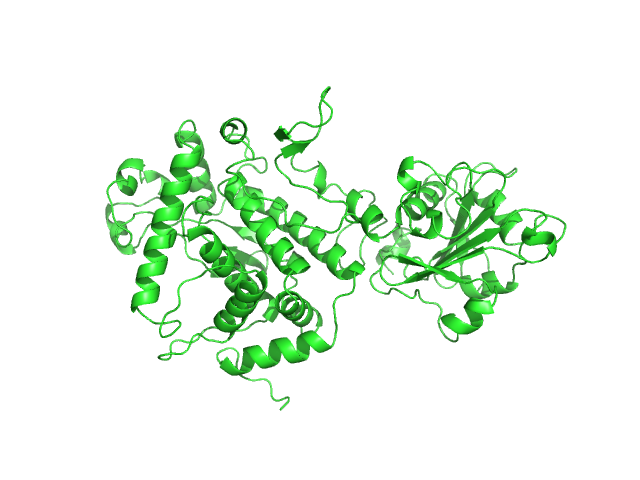
|
| Sample: |
Apoptosis inducing protein monomer, 57 kDa Photobacterium damselae subsp. … protein
|
| Buffer: |
50 mM Hepes, 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Apr 13
|
Unconventional structure and mechanisms for membrane interaction and translocation of the NF-κB-targeting toxin AIP56.
Nat Commun 14(1):7431 (2023)
Lisboa J, Pereira C, Pinto RD, Rodrigues IS, Pereira LMG, Pinheiro B, Oliveira P, Pereira PJB, Azevedo JE, Durand D, Benz R, do Vale A, Dos Santos NMS
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ribosome maturation factor RimP monomer, 18 kDa Staphylococcus aureus (strain … protein
30S ribosomal protein S12 monomer, 15 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
50 mM sodium phosphate, 200 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at Xeuss 3.0 SAXS/WAXS System, JINR on 2023 Feb 16
|
Structural aspects of RimP binding on small ribosomal subunit from Staphylococcus aureus.
Structure (2023)
Garaeva N, Fatkhullin B, Murzakhanov F, Gafurov M, Golubev A, Bikmullin A, Glazyrin M, Kieffer B, Jenner L, Klochkov V, Aganov A, Rogachev A, Ivankov O, Validov S, Yusupov M, Usachev K
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
100 mM sodium acetate, pH 4.6, 2.0 M sodium formate, pH: 4.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 28
|
Dependence of concentration of precursor clusters formed in lysozyme crystallization solutions on degree of supersaturation and its effect on character of solution transition from liquid to condensed phase
Petr Konarev
|
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
200 mM K/Na tartrate, 100 mM tri-sodium citrate pH 5.6, 2.0 M ammonium sulfate, pH: 5.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 28
|
Dependence of concentration of precursor clusters formed in lysozyme crystallization solutions on degree of supersaturation and its effect on character of solution transition from liquid to condensed phase
Petr Konarev
|
|
|
|
|
|
|
|
| Sample: |
DNA repair protein RAD51 homolog 1 (F86E A89E) monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 200 mM Na2SO4, 5% glycerol, 0.1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Apr 7
|
Isolation and characterization of monomeric human RAD51: a novel tool for investigating homologous recombination in cancer
Angewandte Chemie International Edition (2023)
Rinaldi F, Schipani F, Balboni B, Catalano F, Marotta R, Myers S, Previtali V, Veronesi M, Scietti L, Cecatiello V, Pasqualato S, Ortega J, Girotto S, Cavalli A
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA repair protein RAD51 homolog 1 (F86E A89E) monomer, 37 kDa Homo sapiens protein
Breast cancer type 2 susceptibility protein monomer, 4 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 50 mM Na2SO4, 2% v/v sucrose, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Dec 7
|
Isolation and characterization of monomeric human RAD51: a novel tool for investigating homologous recombination in cancer
Angewandte Chemie International Edition (2023)
Rinaldi F, Schipani F, Balboni B, Catalano F, Marotta R, Myers S, Previtali V, Veronesi M, Scietti L, Cecatiello V, Pasqualato S, Ortega J, Girotto S, Cavalli A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
70 |
nm3 |
|
|