|
|
|
|
|
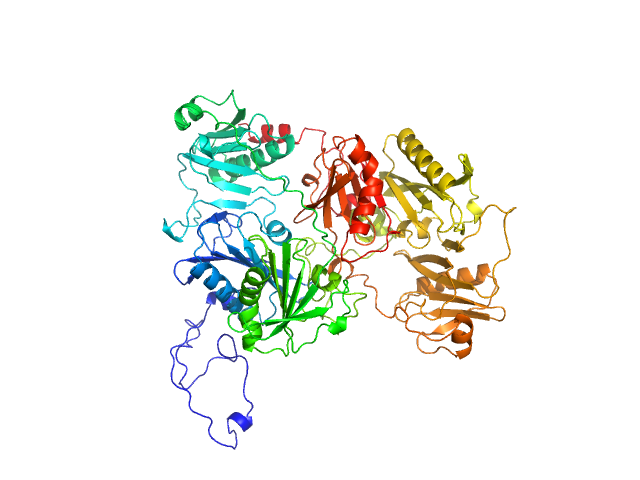
| Sample: |
DNA repair protein RAD51 homolog 1 (F86E, A89E, His-Tagged) monomer, 40 kDa Homo sapiens protein
Breast cancer type 2 susceptibility protein monomer, 6 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 200 mM Na2SO4, 5% glycerol, 0.1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Apr 7
|
Isolation and characterization of monomeric human RAD51: a novel tool for investigating homologous recombination in cancer
Angewandte Chemie International Edition (2023)
Rinaldi F, Schipani F, Balboni B, Catalano F, Marotta R, Myers S, Previtali V, Veronesi M, Scietti L, Cecatiello V, Pasqualato S, Ortega J, Girotto S, Cavalli A
|
| RgGuinier |
3.0 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
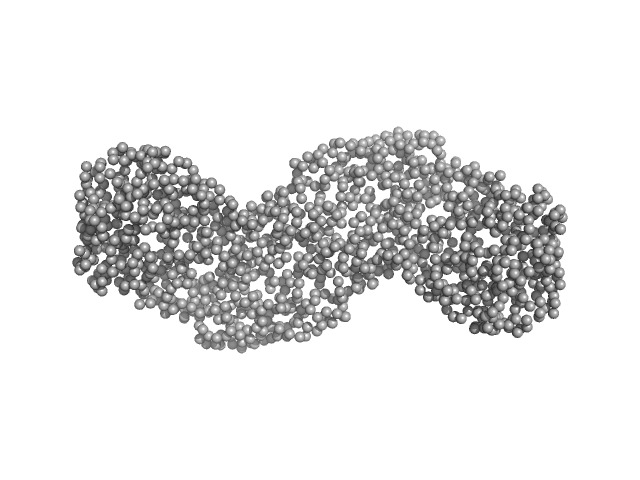
| Sample: |
Gelsolin monomer, 85 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 24
|
Accurate and Efficient SAXS/SANS Implementation Including Solvation Layer Effects Suitable for Molecular Simulations.
J Chem Theory Comput (2023)
Ballabio F, Paissoni C, Bollati M, de Rosa M, Capelli R, Camilloni C
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|
|
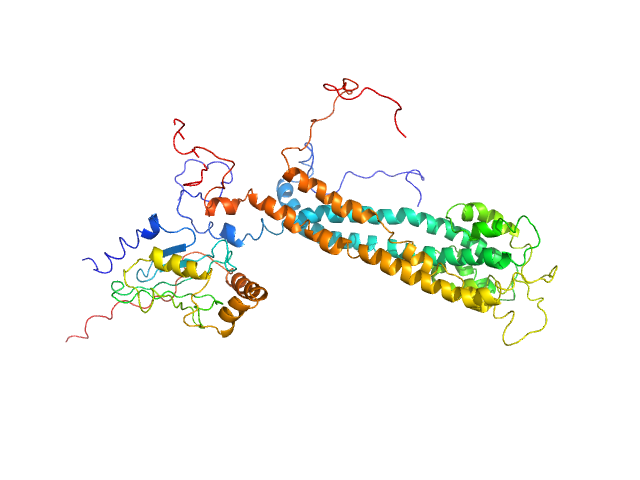
| Sample: |
Gbp6 protein (Gbp7 - Gbp6 protein Mus musculus) dimer, 148 kDa Mus musculus protein
|
| Buffer: |
50 mM Tris, 5 mM MgCl, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2019 Oct 2
|
Integrative modeling of guanylate binding protein dimers
Protein Science (2023)
Schumann W, Loschwitz J, Reiners J, Degrandi D, Legewie L, Stühler K, Pfeffer K, Poschmann G, Smits S, Strodel B
|
| RgGuinier |
5.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
176 |
nm3 |
|
|
|
|
|
|
|
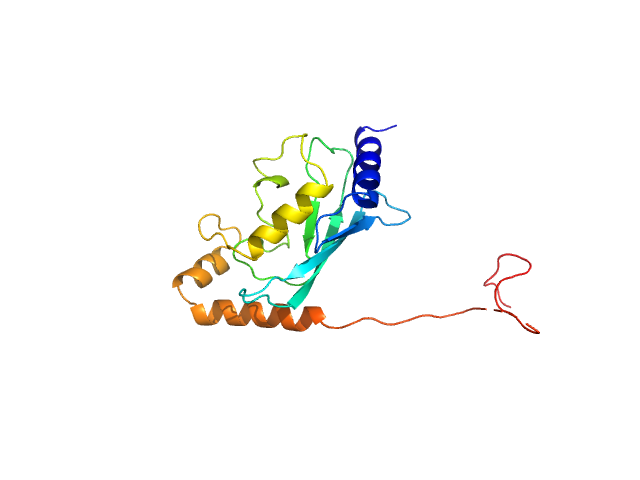
| Sample: |
E3 ubiquitin-protein ligase BRE1 dimer, 50 kDa Saccharomyces cerevisiae (strain … protein
Ubiquitin-conjugating enzyme E2 2 monomer, 20 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Feb 11
|
Structural basis for the role of C-terminus acidic tail of Saccharomyces cerevisiae ubiquitin-conjugating enzyme (Rad6) in E3 ligase (Bre1) mediated recognition of histones
International Journal of Biological Macromolecules :127717 (2023)
Yadav P, Gupta M, Wazahat R, Islam Z, Tsutakawa S, Kamthan M, Kumar P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ubiquitin-conjugating enzyme E2 2 monomer, 20 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Feb 11
|
Structural basis for the role of C-terminus acidic tail of Saccharomyces cerevisiae ubiquitin-conjugating enzyme (Rad6) in E3 ligase (Bre1) mediated recognition of histones
International Journal of Biological Macromolecules :127717 (2023)
Yadav P, Gupta M, Wazahat R, Islam Z, Tsutakawa S, Kamthan M, Kumar P
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 150 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
5.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 150 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
5.3 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
300 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 150 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
5.4 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
325 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 150 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
5.5 |
nm |
| Dmax |
21.4 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Contactin-2 dimer, 150 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
6.0 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
380 |
nm3 |
|
|