|
|
|
|

|
| Sample: |
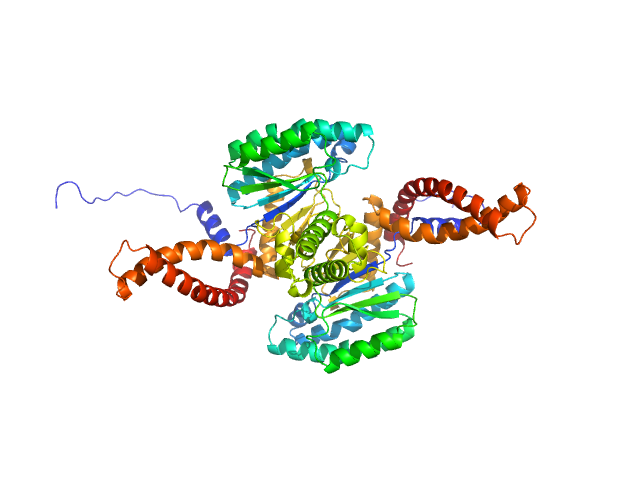
Dystrophin (R11-15 human dystrophin fragment) monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% v/v D2O, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 1
|
How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids.
J Struct Biol :107411 (2019)
Mias-Lucquin D, Dos Santos Morais R, Chéron A, Lagarrigue M, Winder SJ, Chenuel T, Pérez J, Appavou MS, Martel A, Alviset G, Le Rumeur E, Combet S, Hubert JF, Delalande O
|
| RgGuinier |
8.1 |
nm |
| Dmax |
29.6 |
nm |
| VolumePorod |
231 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dehydrodolichyl diphosphate synthase complex subunit DHDDS dimer, 78 kDa Homo sapiens protein
|
| Buffer: |
Tris-HCl, 150 mM NaCl, 20 mM 2-mercaptoethanol and 0.02% Triton-X100, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 2
|
Structural Characterization of Full-Length Human Dehydrodolichyl Diphosphate Synthase Using an Integrative Computational and Experimental Approach.
Biomolecules 9(11) (2019)
Lisnyansky Bar-El M, Lee SY, Ki AY, Kapelushnik N, Loewenstein A, Chung KY, Schneidman-Duhovny D, Giladi M, Newman H, Haitin Y
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
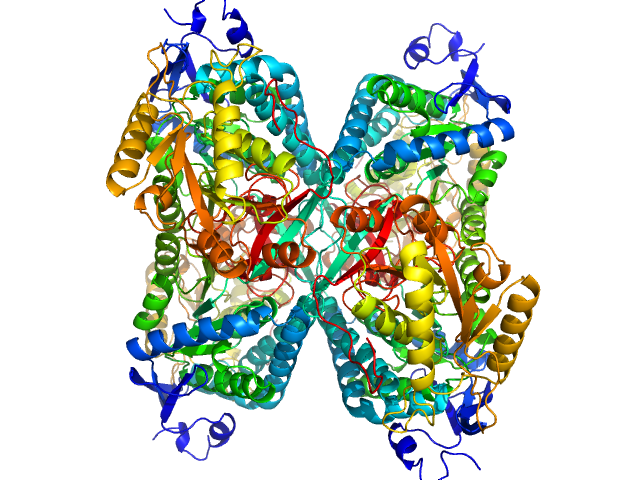
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
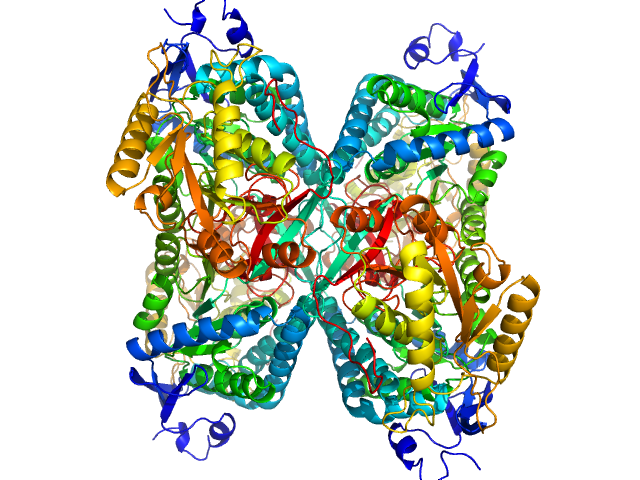
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
326 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
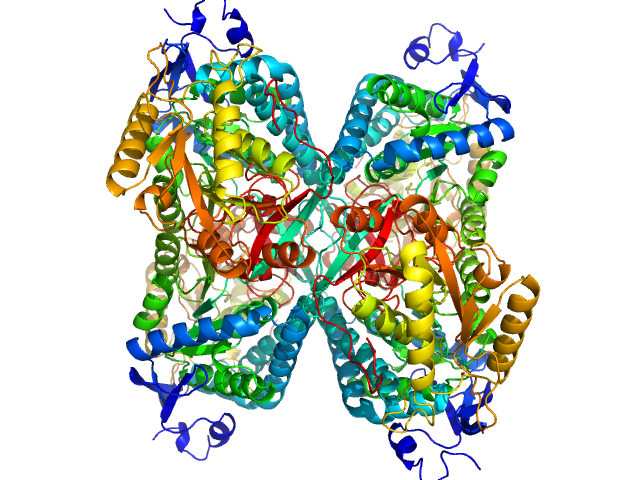
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
315 |
nm3 |
|
|
|
|
|
|
|
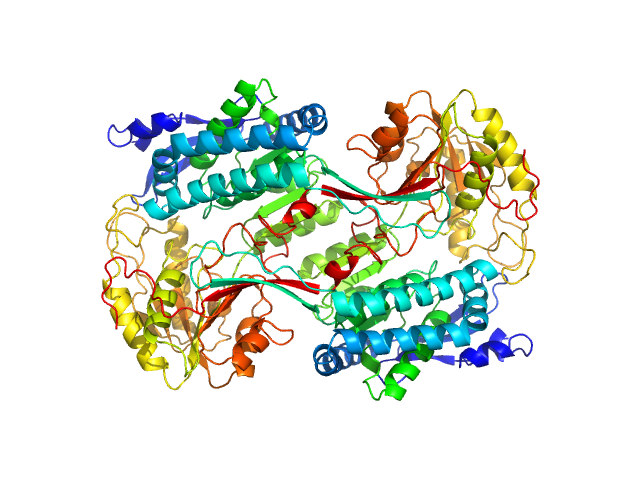
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399Q, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
237 |
nm3 |
|
|
|
|
|
|
|
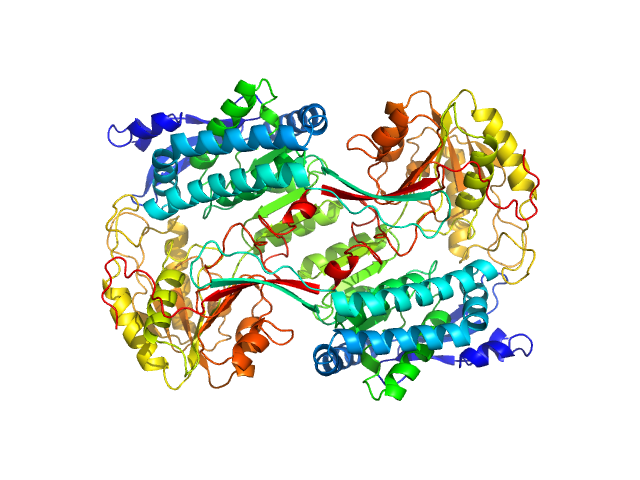
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399Q, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
|
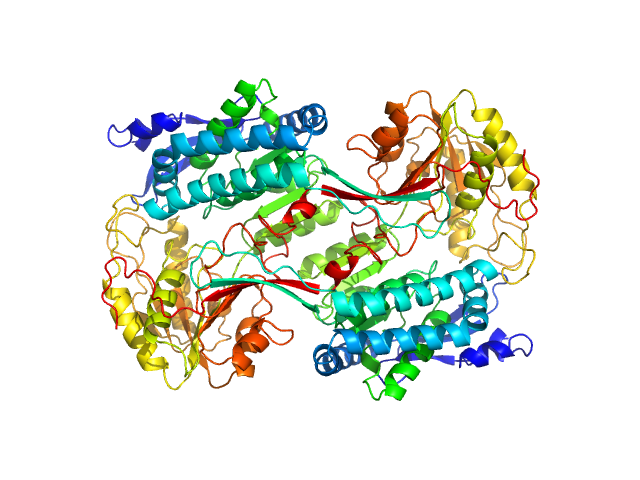
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399Q, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
255 |
nm3 |
|
|
|
|
|
|
|
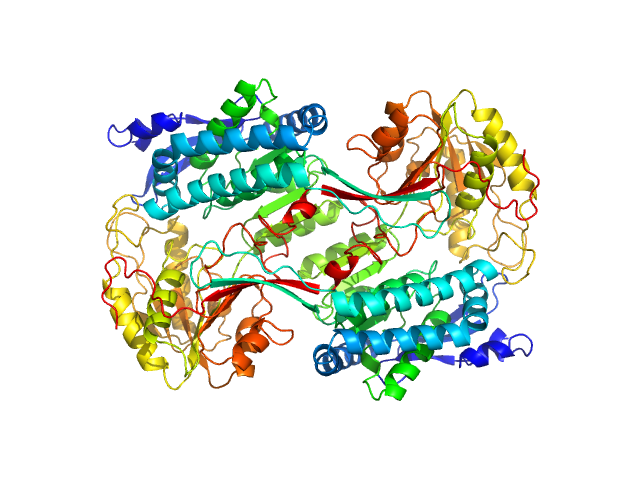
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
256 |
nm3 |
|
|
|
|
|
|
|
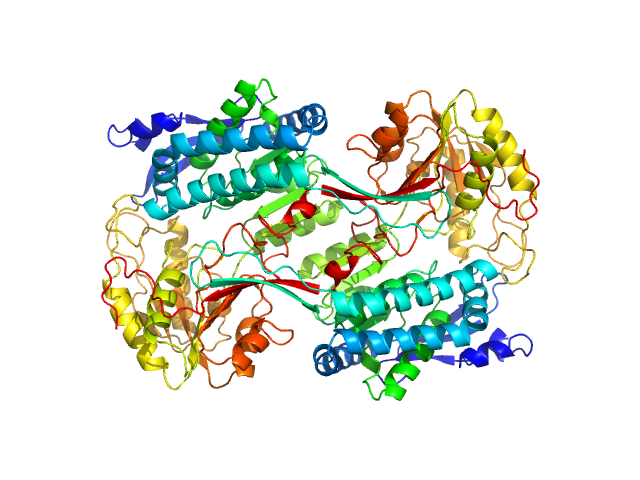
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
260 |
nm3 |
|
|