|
|
|
|
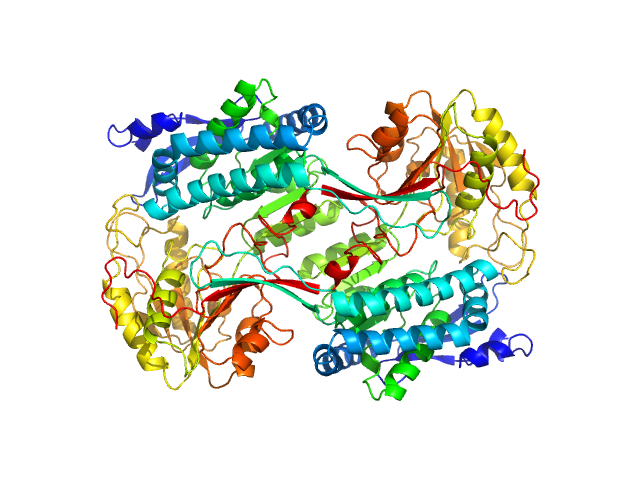
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399D, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.9 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
272 |
nm3 |
|
|
|
|
|
|
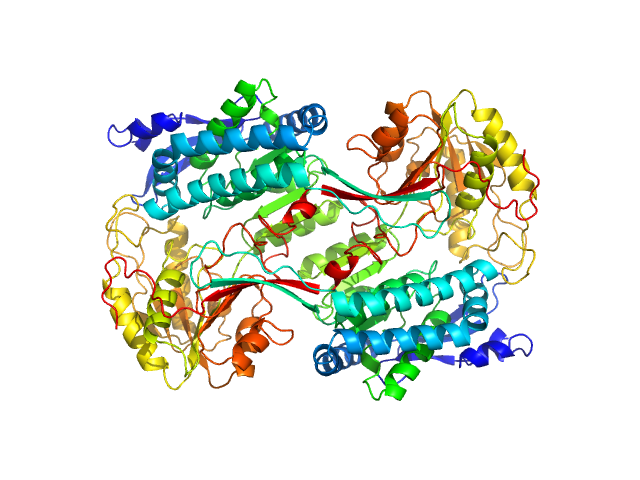
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399G, 55 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
|
|
|
|
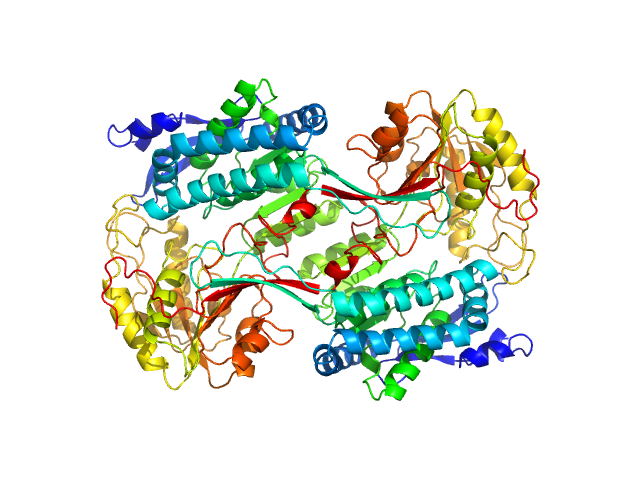
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399G, 55 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
250 |
nm3 |
|
|
|
|
|
|
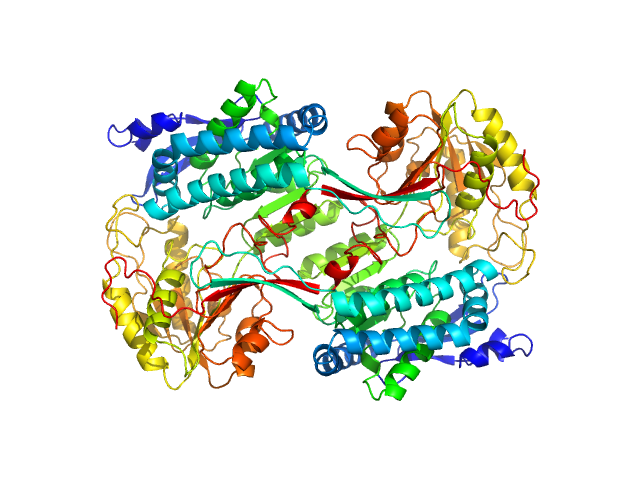
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399G, 55 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aryl-hydrocarbon-interacting protein-like 1(1-316) monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 2.5 % glycerol and 6 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Jul 17
|
Interaction of the tetratricopeptide repeat domain of aryl hydrocarbon receptor-interacting protein-like 1 with the regulatory Pγ subunit of phosphodiesterase 6.
J Biol Chem 294(43):15795-15807 (2019)
Yadav RP, Boyd K, Yu L, Artemyev NO
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Adhesion G-protein coupled receptor G6 S2 monomer, 88 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Jun 28
|
Structural basis for adhesion G protein-coupled receptor Gpr126 function
Nature Communications 11(1) (2020)
Leon K, Cunningham R, Riback J, Feldman E, Li J, Sosnick T, Zhao M, Monk K, Araç D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
199 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Adhesion G-protein coupled receptor G6 S1 monomer, 91 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Jun 28
|
Structural basis for adhesion G protein-coupled receptor Gpr126 function
Nature Communications 11(1) (2020)
Leon K, Cunningham R, Riback J, Feldman E, Li J, Sosnick T, Zhao M, Monk K, Araç D
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cytohesin-2; ARNO truncation mutant monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Nov 25
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
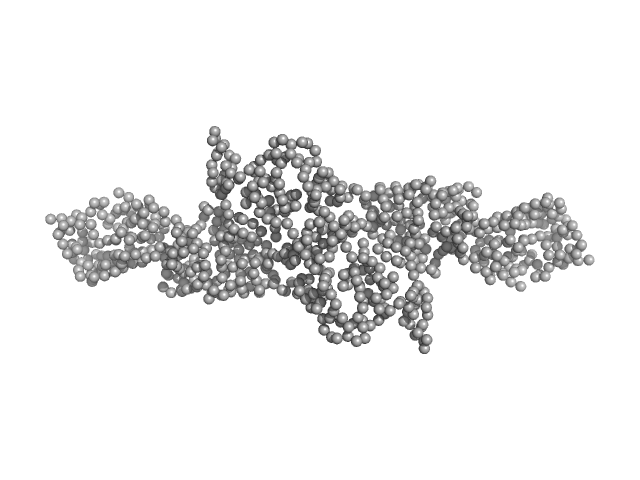
|
| Sample: |
Cytohesin-2 ARF nucleotide-binding site opener dimer, 93 kDa Homo sapiens protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 23
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.7 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mothers against decapentaplegic homolog 2 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jul 2
|
Structural basis for distinct roles of SMAD2 and SMAD3 in FOXH1 pioneer-directed TGF-β signaling.
Genes Dev 33(21-22):1506-1524 (2019)
Aragón E, Wang Q, Zou Y, Morgani SM, Ruiz L, Kaczmarska Z, Su J, Torner C, Tian L, Hu J, Shu W, Agrawal S, Gomes T, Márquez JA, Hadjantonakis AK, Macias MJ, Massagué J
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
35 |
nm3 |
|
|