|
|
|
|
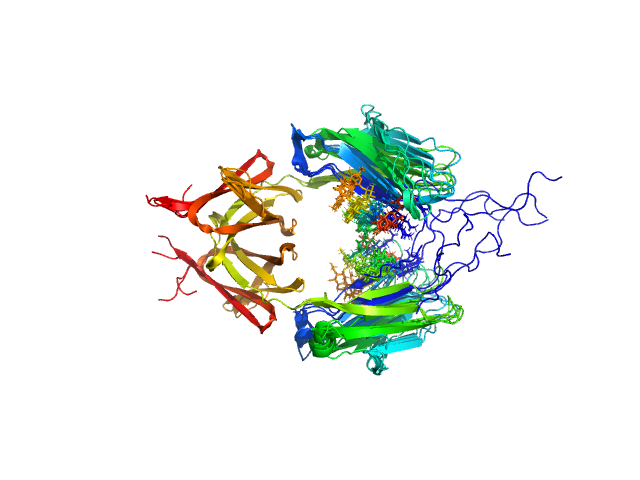
|
| Sample: |
Immunoglobulin heavy constant gamma 2 dimer, 52 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
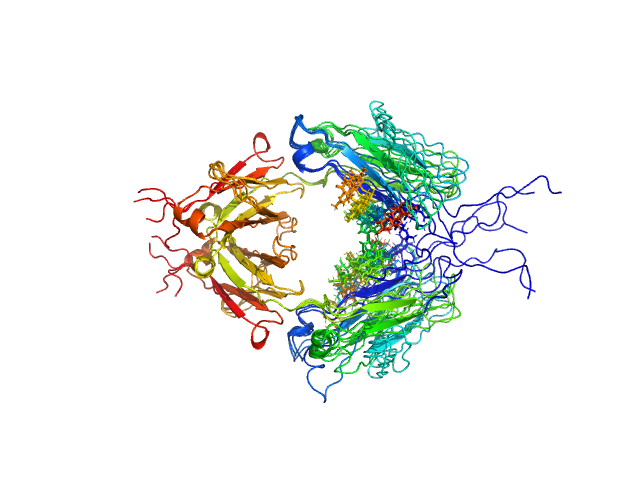
|
| Sample: |
Immunoglobulin heavy constant gamma 1 M255Y/S257T/T259E dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human telomerase RNA template component, DNA counterpart, nucleotides 1-20 dimer, 13 kDa Homo sapiens DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl,, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge.
Nucleic Acids Res 46(10):5319-5331 (2018)
Meier M, Moya-Torres A, Krahn NJ, McDougall MD, Orriss GL, McRae EKS, Booy EP, McEleney K, Patel TR, McKenna SA, Stetefeld J
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
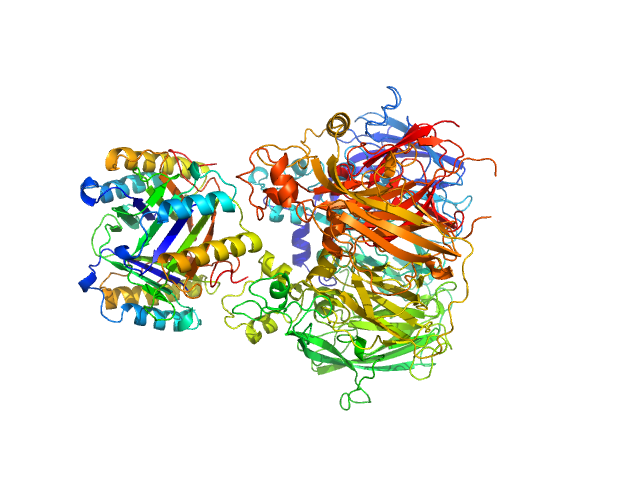
|
| Sample: |
Macrophage migration inhibitory factor trimer, 37 kDa Homo sapiens protein
Ceruloplasmin monomer, 122 kDa Homo sapiens protein
|
| Buffer: |
50mkm CuSO4, 100mM Hepes, pH: 7.4 |
| Experiment: |
SAXS
data collected at HECUS System-3, None on 2015 Dec 22
|
Structural Study of the Complex Formed by Ceruloplasmin and Macrophage Migration Inhibitory Factor.
Biochemistry (Mosc) 83(6):701-707 (2018)
Sokolov AV, Dadinova LA, Petoukhov MV, Bourenkov G, Dubova KM, Amarantov SV, Volkov VV, Kostevich VA, Gorbunov NP, Grudinina NA, Vasilyev VB, Samygina VR
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
228 |
nm3 |
|
|
|
|
|
|
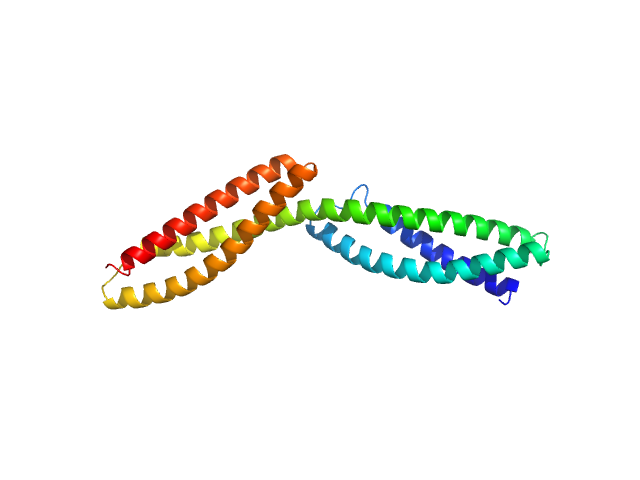
|
| Sample: |
Dystrophin central domain repeats 1 to 2 monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Oct 7
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
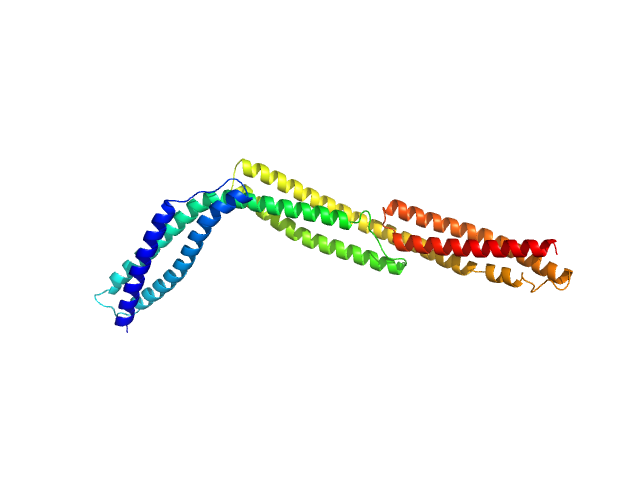
|
| Sample: |
Dystrophin central domain repeats 1 to 3 monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Sep 9
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
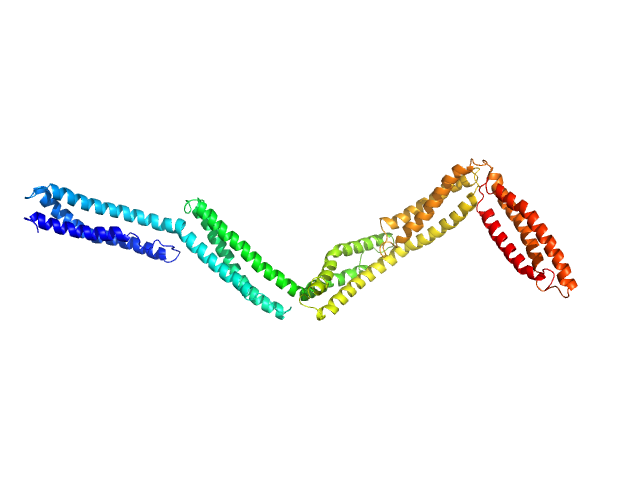
|
| Sample: |
Dystrophin central domain repeats 11 to 15. monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2011 Apr 11
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
5.8 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dystrophin central domain repeats 4 to 9 monomer, 76 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Sep 19
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
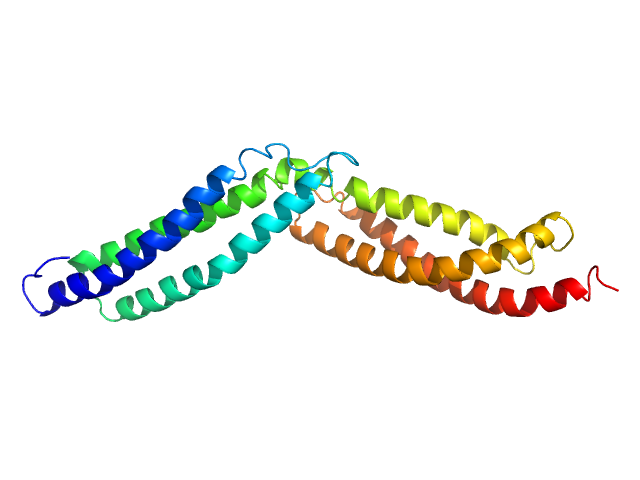
|
| Sample: |
Dystrophin central domain repeats 16 to 17. monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Sep 19
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
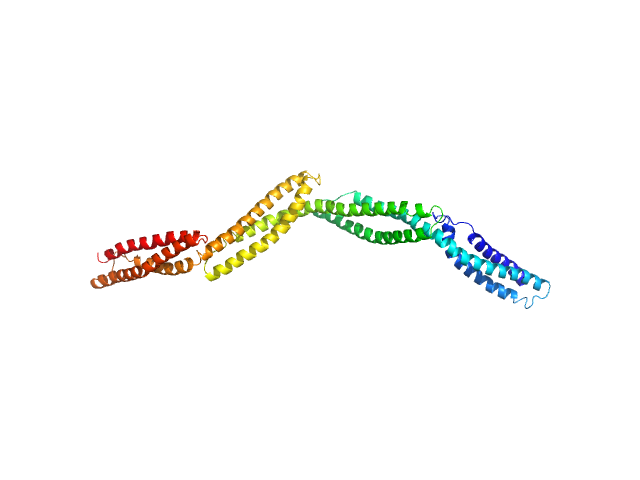
|
| Sample: |
Dystrophin central domain repeats 16 to 19. monomer, 50 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol 5% acetonitrile, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Oct 4
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
4.6 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|