|
|
|
|
|
| Sample: |
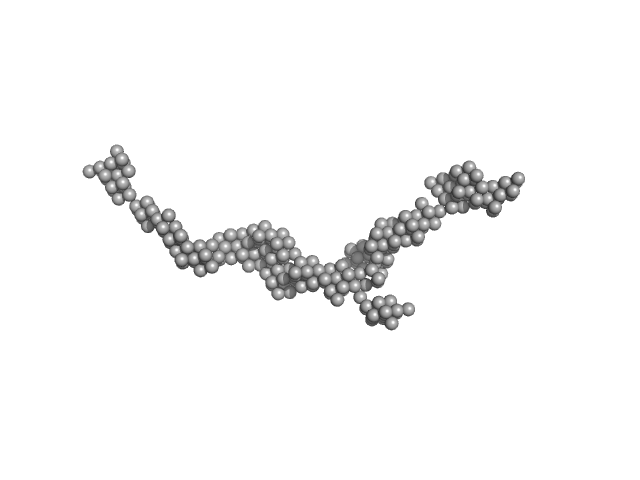
Myomesin-1 dimer, 121 kDa Homo sapiens protein
|
| Buffer: |
25 mMTris-HCl, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 10
|
Superhelical architecture of the myosin filament-linking protein myomesin with unusual elastic properties.
PLoS Biol 10(2):e1001261 (2012)
Pinotsis N, Chatziefthimiou SD, Berkemeier F, Beuron F, Mavridis IM, Konarev PV, Svergun DI, Morris E, Rief M, Wilmanns M
|
| RgGuinier |
9.6 |
nm |
| Dmax |
37.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
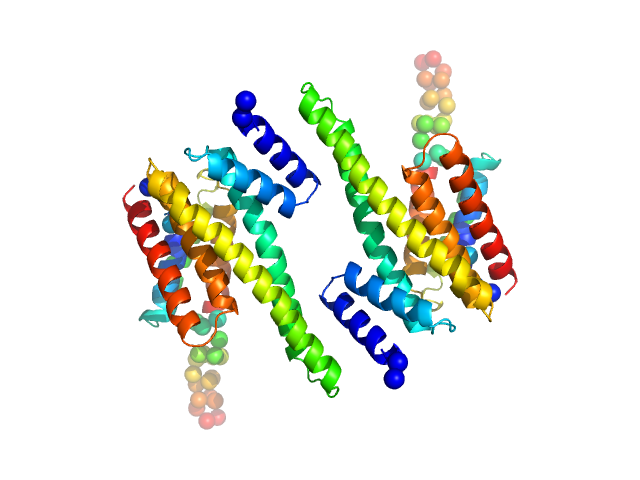
14-3-3 protein beta/alpha dimer, 58 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2011 Apr 7
|
The weak complex between RhoGAP protein ARHGAP22 and signal regulatory protein 14-3-3 has 1:2 stoichiometry and a single peptide binding mode.
PLoS One 7(8):e41731 (2012)
Hu SH, Whitten AE, King GJ, Jones A, Rowland AF, James DE, Martin JL
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
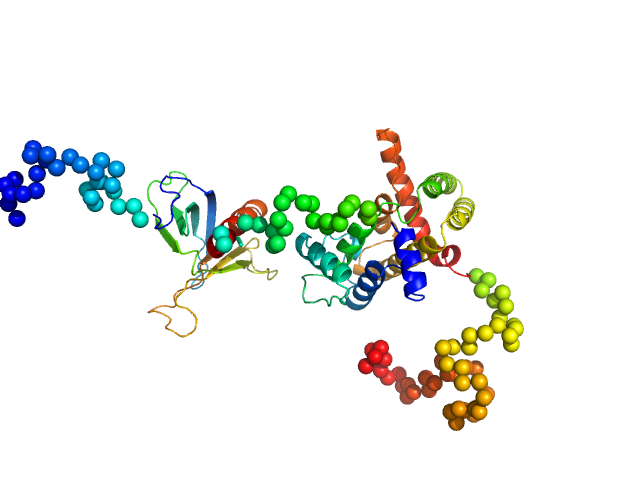
Rho GTPase-activating protein 22 monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2011 Apr 7
|
The weak complex between RhoGAP protein ARHGAP22 and signal regulatory protein 14-3-3 has 1:2 stoichiometry and a single peptide binding mode.
PLoS One 7(8):e41731 (2012)
Hu SH, Whitten AE, King GJ, Jones A, Rowland AF, James DE, Martin JL
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
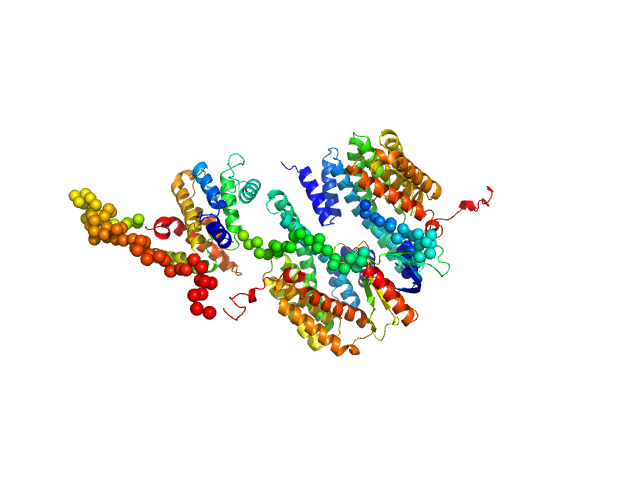
14-3-3 protein beta/alpha dimer, 58 kDa Homo sapiens protein
Rho GTPase-activating protein 22 monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2011 Apr 7
|
The weak complex between RhoGAP protein ARHGAP22 and signal regulatory protein 14-3-3 has 1:2 stoichiometry and a single peptide binding mode.
PLoS One 7(8):e41731 (2012)
Hu SH, Whitten AE, King GJ, Jones A, Rowland AF, James DE, Martin JL
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
195 |
nm3 |
|
|
|
|
|
|
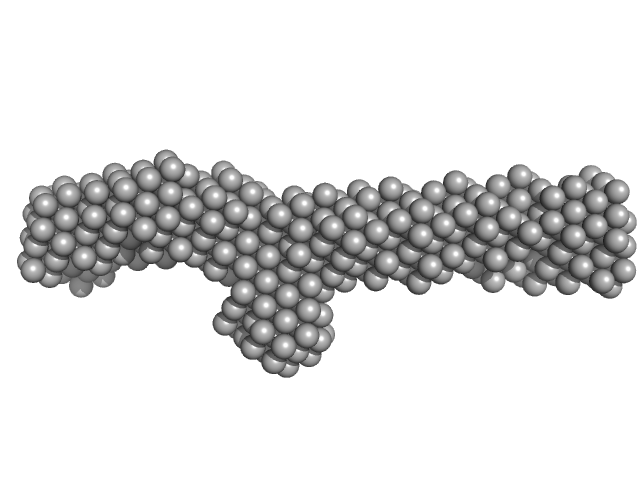
|
| Sample: |
Plectin monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 2.5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at cSAXS, Swiss Light Source on 2009 Jul 24
|
The structure of the plakin domain of plectin reveals a non-canonical SH3 domain interacting with its fourth spectrin repeat.
J Biol Chem 286(14):12429-38 (2011)
Ortega E, Buey RM, Sonnenberg A, de Pereda JM
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
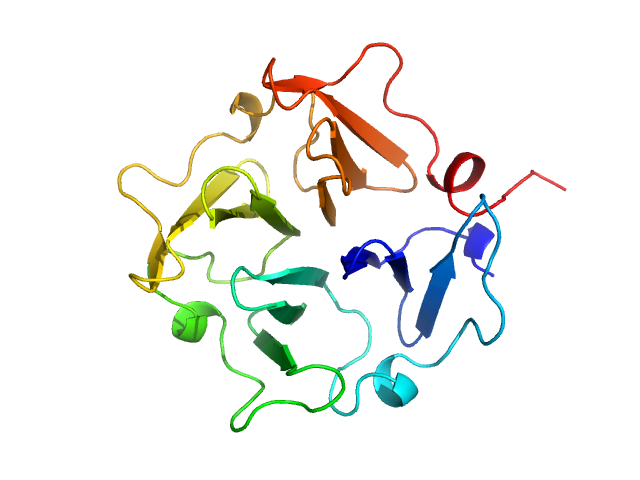
|
| Sample: |
Hemopexin monomer, 23 kDa Homo sapiens protein
Hemopexin dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 10 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 25
|
The Dimer Interface of the Membrane Type 1 Matrix Metalloproteinase Hemopexin Domain
Journal of Biological Chemistry 286(9):7587-7600 (2011)
Tochowicz A, Goettig P, Evans R, Visse R, Shitomi Y, Palmisano R, Ito N, Richter K, Maskos K, Franke D, Svergun D, Nagase H, Bode W, Itoh Y
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
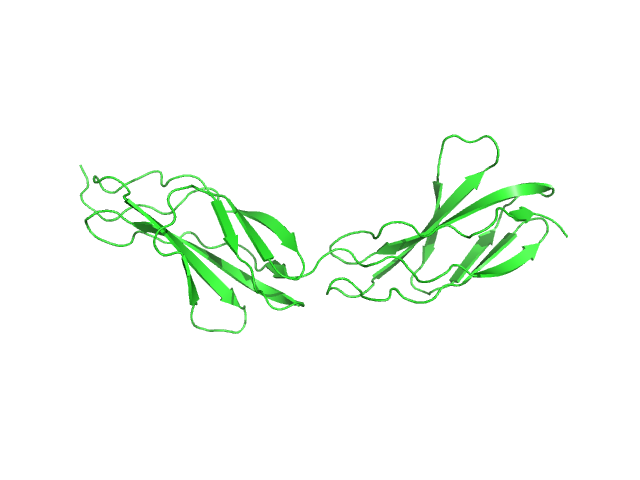
|
| Sample: |
Titin monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jul 3
|
The Structure of the FnIII Tandem A77-A78 Points to a Periodically Conserved Architecture in the Myosin-Binding Region of Titin
Journal of Molecular Biology 401(5):843-853 (2010)
Bucher R, Svergun D, Muhle-Goll C, Mayans O
|
| RgGuinier |
2.5 |
nm |
| Dmax |
90.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
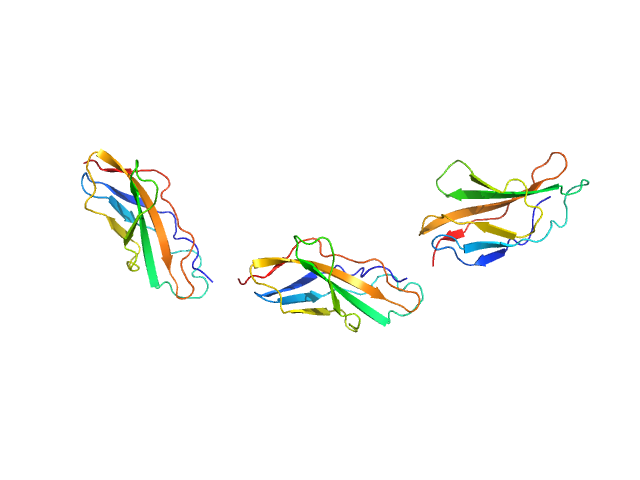
|
| Sample: |
Titin monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jul 3
|
The Structure of the FnIII Tandem A77-A78 Points to a Periodically Conserved Architecture in the Myosin-Binding Region of Titin
Journal of Molecular Biology 401(5):843-853 (2010)
Bucher R, Svergun D, Muhle-Goll C, Mayans O
|
| RgGuinier |
3.7 |
nm |
| Dmax |
130.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Titin monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Oct 6
|
The Structure of the FnIII Tandem A77-A78 Points to a Periodically Conserved Architecture in the Myosin-Binding Region of Titin
Journal of Molecular Biology 401(5):843-853 (2010)
Bucher R, Svergun D, Muhle-Goll C, Mayans O
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Factor H CCP modules 10 to 15 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM Potassium Phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Dec 2
|
The central portion of factor H (modules 10-15) is compact and contains a structurally deviant CCP module.
J Mol Biol 395(1):105-22 (2010)
Schmidt CQ, Herbert AP, Mertens HD, Guariento M, Soares DC, Uhrin D, Rowe AJ, Svergun DI, Barlow PN
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
68 |
nm3 |
|
|