|
|
|
|
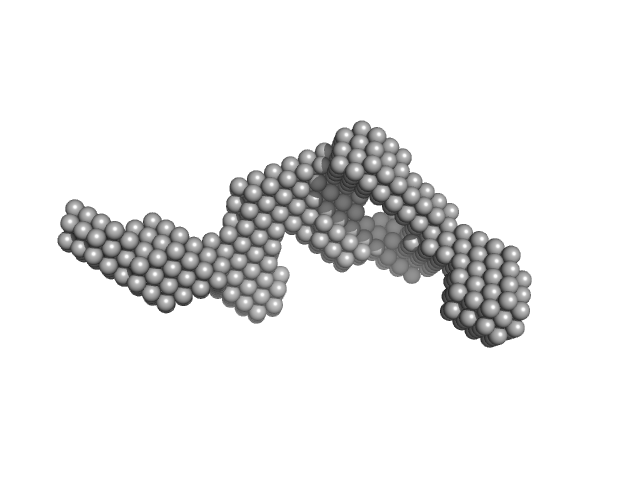
|
| Sample: |
Factor H CCP modules 11 to 14 monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
50 mM Potassium Phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Dec 2
|
The central portion of factor H (modules 10-15) is compact and contains a structurally deviant CCP module.
J Mol Biol 395(1):105-22 (2010)
Schmidt CQ, Herbert AP, Mertens HD, Guariento M, Soares DC, Uhrin D, Rowe AJ, Svergun DI, Barlow PN
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
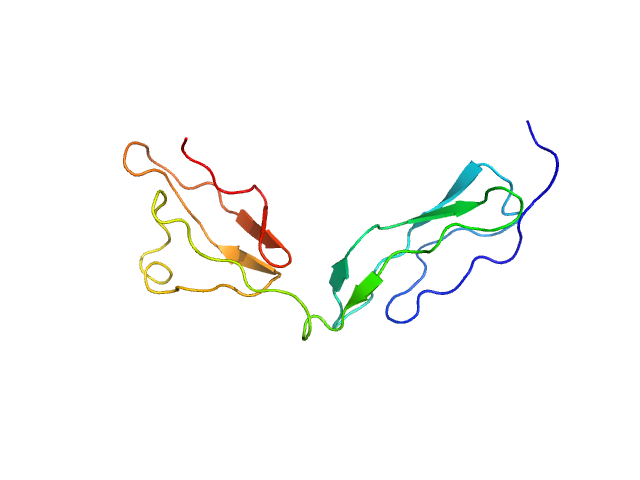
|
| Sample: |
Factor H CCP modules 12 to 13 monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
50 mM Potassium Phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Dec 2
|
The central portion of factor H (modules 10-15) is compact and contains a structurally deviant CCP module.
J Mol Biol 395(1):105-22 (2010)
Schmidt CQ, Herbert AP, Mertens HD, Guariento M, Soares DC, Uhrin D, Rowe AJ, Svergun DI, Barlow PN
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
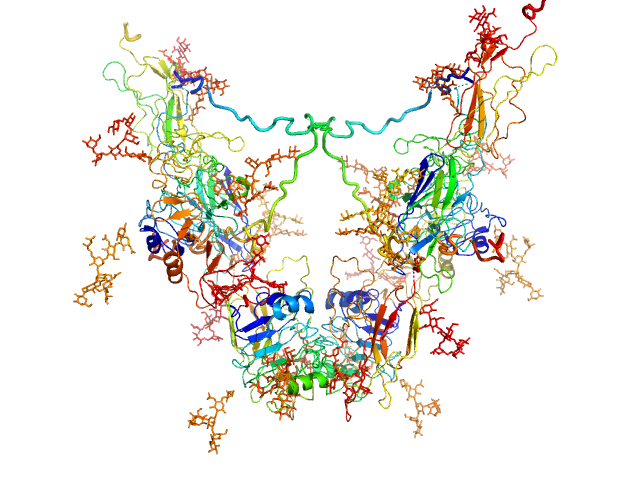
|
| Sample: |
Insulin-like growth factor 1 receptor dimer, 202 kDa Homo sapiens protein
|
| Buffer: |
30 mM Tris, 140 mM NaCl, 0.02% w/v sodium azide,, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, Australian Nuclear Science and Technology Organisation on 2008 Apr 22
|
Solution structure of ectodomains of the insulin receptor family: the ectodomain of the type 1 insulin-like growth factor receptor displays asymmetry of ligand binding accompanied by limited conformational change.
J Mol Biol 394(5):878-92 (2009)
Whitten AE, Smith BJ, Menting JG, Margetts MB, McKern NM, Lovrecz GO, Adams TE, Richards K, Bentley JD, Trewhella J, Ward CW, Lawrence MC
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
357 |
nm3 |
|
|
|
|
|
|
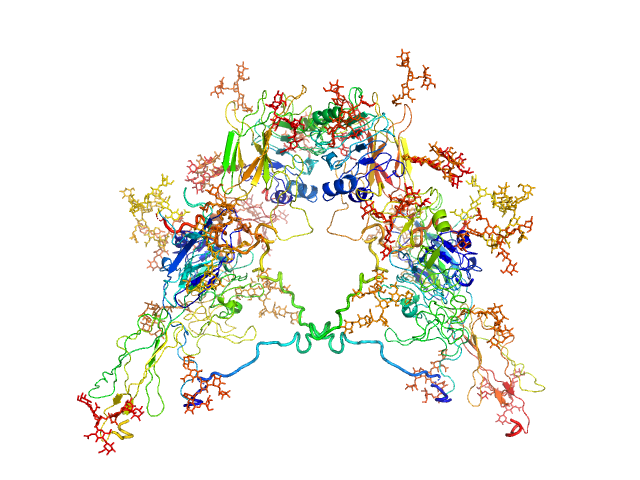
|
| Sample: |
Insulin receptor dimer, 203 kDa Homo sapiens protein
|
| Buffer: |
30 mM Tris, 140 mM NaCl, 0.02% w/v sodium azide,, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, Australian Nuclear Science and Technology Organisation on 2008 Dec 7
|
Solution structure of ectodomains of the insulin receptor family: the ectodomain of the type 1 insulin-like growth factor receptor displays asymmetry of ligand binding accompanied by limited conformational change.
J Mol Biol 394(5):878-92 (2009)
Whitten AE, Smith BJ, Menting JG, Margetts MB, McKern NM, Lovrecz GO, Adams TE, Richards K, Bentley JD, Trewhella J, Ward CW, Lawrence MC
|
| RgGuinier |
5.5 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
385 |
nm3 |
|
|
|
|
|
|
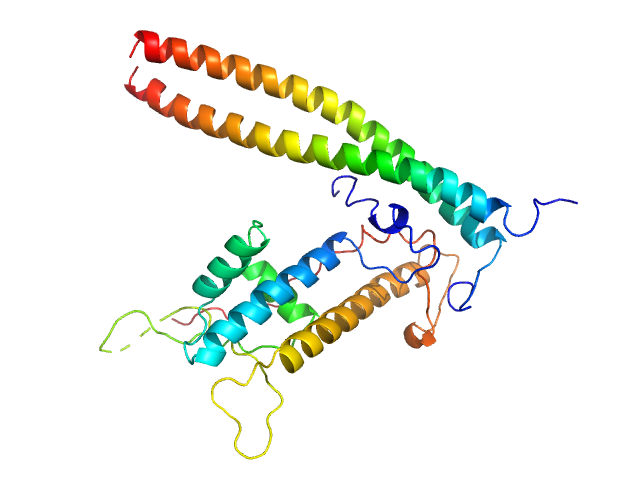
|
| Sample: |
Geminin dimer, 47 kDa Homo sapiens protein
DNA replication factor Cdt1 monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris75 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Aug 13
|
Quaternary structure of the human Cdt1-Geminin complex regulates DNA replication licensing.
Proc Natl Acad Sci U S A 106(47):19807-12 (2009)
De Marco V, Gillespie PJ, Li A, Karantzelis N, Christodoulou E, Klompmaker R, van Gerwen S, Fish A, Petoukhov MV, Iliou MS, Lygerou Z, Medema RH, Blow JJ, Svergun DI, Taraviras S, Perrakis A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
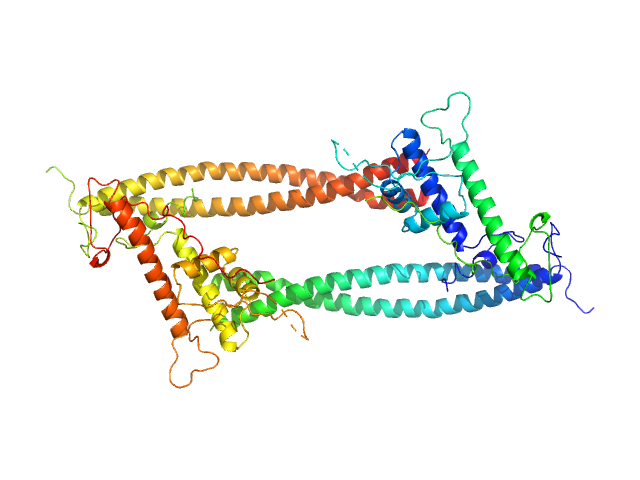
|
| Sample: |
Geminin dimer, 47 kDa Homo sapiens protein
DNA replication factor Cdt1 monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris75 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Oct 5
|
Quaternary structure of the human Cdt1-Geminin complex regulates DNA replication licensing.
Proc Natl Acad Sci U S A 106(47):19807-12 (2009)
De Marco V, Gillespie PJ, Li A, Karantzelis N, Christodoulou E, Klompmaker R, van Gerwen S, Fish A, Petoukhov MV, Iliou MS, Lygerou Z, Medema RH, Blow JJ, Svergun DI, Taraviras S, Perrakis A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DH-PH module of PDZRhoGEF monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Oct 30
|
The solution structure and dynamics of the DH-PH module of PDZRhoGEF in isolation and in complex with nucleotide-free RhoA.
Protein Sci 18(10):2067-79 (2009)
Cierpicki T, Bielnicki J, Zheng M, Gruszczyk J, Kasterka M, Petoukhov M, Zhang A, Fernandez EJ, Svergun DI, Derewenda U, Bushweller JH, Derewenda ZS
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
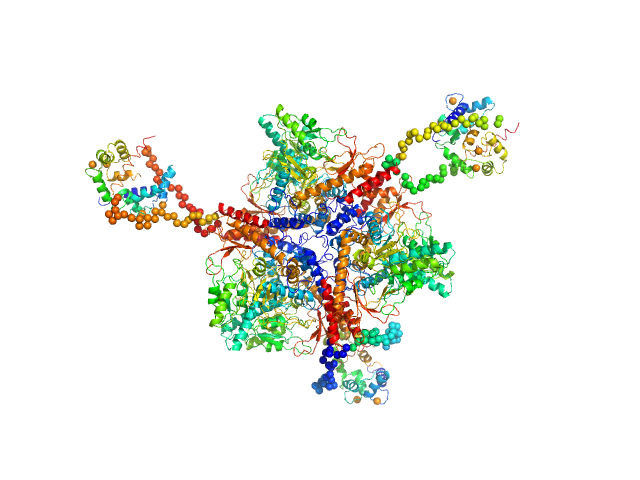
|
| Sample: |
Calmodulin monomer, 17 kDa Homo sapiens protein
Glutamate decarboxylase hexamer, 340 kDa Petunia x hybrida protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Nov 9
|
A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase.
J Mol Biol 392(2):334-51 (2009)
Gut H, Dominici P, Pilati S, Astegno A, Petoukhov MV, Svergun DI, Grütter MG, Capitani G
|
| RgGuinier |
5.9 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
630 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peroxisomal targeting signal 1 receptor (C -terminal) monomer, 48 kDa Homo sapiens protein
Peroxisomal membrane protein PEX14 (N-terminal) monomer, 7 kDa Homo sapiens protein
PTS1-BP monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH (pH 7.5), 100mM KCl, and 20mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Oct 3
|
Solution structure of human Pex5.Pex14.PTS1 protein complexes obtained by small angle X-ray scattering.
J Biol Chem 284(37):25334-42 (2009)
Shiozawa K, Konarev PV, Neufeld C, Wilmanns M, Svergun DI
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Peroxisomal targeting signal 1 receptor monomer, 71 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH (pH 7.5), 100mM KCl, and 20mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 24
|
Solution structure of human Pex5.Pex14.PTS1 protein complexes obtained by small angle X-ray scattering.
J Biol Chem 284(37):25334-42 (2009)
Shiozawa K, Konarev PV, Neufeld C, Wilmanns M, Svergun DI
|
| RgGuinier |
5.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
181 |
nm3 |
|
|