|
|
|
|
|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Synthetic peptide AE234 monomer, 1 kDa protein
|
| Buffer: |
25 mM Sodium Phosphate 5 % Glycerol 50 mM NaSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jun 10
|
A flexible multidomain structure drives the function of the urokinase-type plasminogen activator receptor (uPAR).
J Biol Chem 287(41):34304-15 (2012)
Mertens HD, Kjaergaard M, Mysling S, Gårdsvoll H, Jørgensen TJ, Svergun DI, Ploug M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Urokinase-type plasminogen activator monomer, 49 kDa Homo sapiens protein
|
| Buffer: |
25 mM Sodium Phosphate 5 % Glycerol 50 mM NaSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jun 10
|
A flexible multidomain structure drives the function of the urokinase-type plasminogen activator receptor (uPAR).
J Biol Chem 287(41):34304-15 (2012)
Mertens HD, Kjaergaard M, Mysling S, Gårdsvoll H, Jørgensen TJ, Svergun DI, Ploug M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human Chromatin Remodeler CHD4 (363-1353) monomer, 117 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 5% Glycerol 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Nov 13
|
The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4.
J Mol Biol 422(1):3-17 (2012)
Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, Du X, Bartke T, Zhang W, Edlich C, Berridge G, Chen Y, Burgess-Brown NA, Kouzarides T, Wiechens N, Owen-Hughes T, Svergun DI, Gileadi O, Laue ED
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.4 |
nm |
|
|
|
|
|
|

|
| Sample: |
Human Chromatin Remodeler CHD4 (494-1353) monomer, 101 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Nov 13
|
The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4.
J Mol Biol 422(1):3-17 (2012)
Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, Du X, Bartke T, Zhang W, Edlich C, Berridge G, Chen Y, Burgess-Brown NA, Kouzarides T, Wiechens N, Owen-Hughes T, Svergun DI, Gileadi O, Laue ED
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Human Chromatin Remodeler CHD4 (685-1233) monomer, 63 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Nov 13
|
The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4.
J Mol Biol 422(1):3-17 (2012)
Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, Du X, Bartke T, Zhang W, Edlich C, Berridge G, Chen Y, Burgess-Brown NA, Kouzarides T, Wiechens N, Owen-Hughes T, Svergun DI, Gileadi O, Laue ED
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Human Chromatin Remodeler CHD4 (363-682) monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Nov 13
|
The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4.
J Mol Biol 422(1):3-17 (2012)
Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, Du X, Bartke T, Zhang W, Edlich C, Berridge G, Chen Y, Burgess-Brown NA, Kouzarides T, Wiechens N, Owen-Hughes T, Svergun DI, Gileadi O, Laue ED
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
CYNEX4 FRET probe, (eYFP-AnnexinA4-eCFP) monomer, 93 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1 mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Sep 11
|
Conformational analysis of a genetically encoded FRET biosensor by SAXS.
Biophys J 102(12):2866-75 (2012)
Mertens HD, Piljić A, Schultz C, Svergun DI
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
135 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
CYNEX4 FRET probe, (eYFP-AnnexinA4-eCFP) T266D mutant monomer, 93 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Sep 11
|
Conformational analysis of a genetically encoded FRET biosensor by SAXS.
Biophys J 102(12):2866-75 (2012)
Mertens HD, Piljić A, Schultz C, Svergun DI
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
146 |
nm3 |
|
|
|
|
|
|
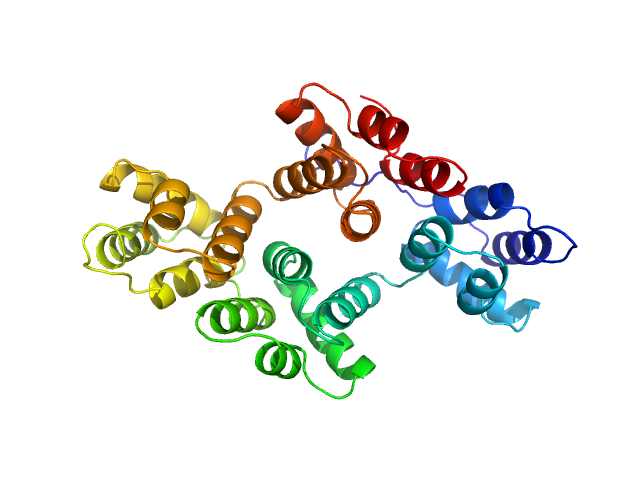
|
| Sample: |
Annexin-A4 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Sep 11
|
Conformational analysis of a genetically encoded FRET biosensor by SAXS.
Biophys J 102(12):2866-75 (2012)
Mertens HD, Piljić A, Schultz C, Svergun DI
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polyglutamine-binding protein 1 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1mM DTT,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 18
|
Solution model of the intrinsically disordered polyglutamine tract-binding protein-1.
Biophys J 102(7):1608-16 (2012)
Rees M, Gorba C, de Chiara C, Bui TT, Garcia-Maya M, Drake AF, Okazawa H, Pastore A, Svergun D, Chen YW
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
51 |
nm3 |
|
|