|
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
3.3 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Nov 15
|
Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
BMC Biol 20(1):176 (2022)
Léger C, Pitard I, Sadi M, Carvalho N, Brier S, Mechaly A, Raoux-Barbot D, Davi M, Hoos S, Weber P, Vachette P, Durand D, Haouz A, Guijarro JI, Ladant D, Chenal A
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Calmidazolium monomer, 1 kDa
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Nov 15
|
Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
BMC Biol 20(1):176 (2022)
Léger C, Pitard I, Sadi M, Carvalho N, Brier S, Mechaly A, Raoux-Barbot D, Davi M, Hoos S, Weber P, Vachette P, Durand D, Haouz A, Guijarro JI, Ladant D, Chenal A
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Thyroglobulin dimer, 610 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, 150mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2020 Jul 17
|
Thyroglobulin - human and bovine SAXS/WAXS data
James Byrnes
|
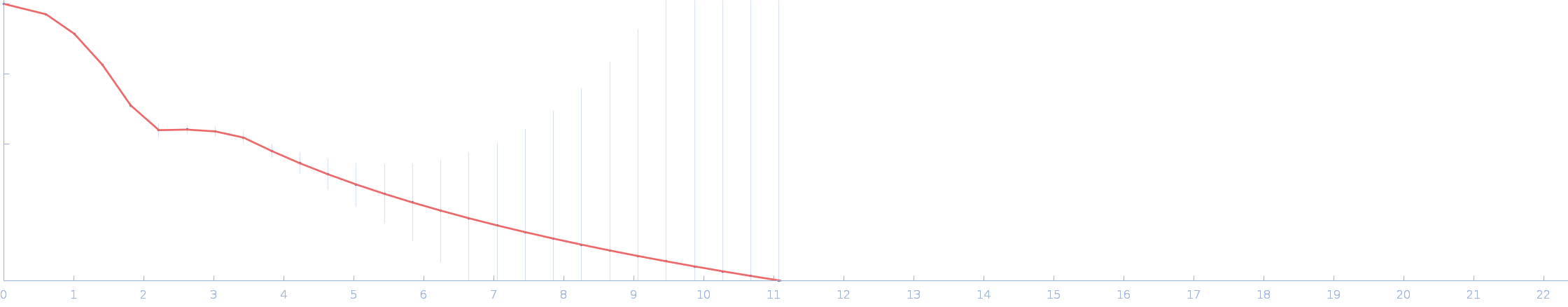
| RgGuinier |
7.7 |
nm |
| Dmax |
26.3 |
nm |
| VolumePorod |
1800 |
nm3 |
|
|
|
|
|
|
|
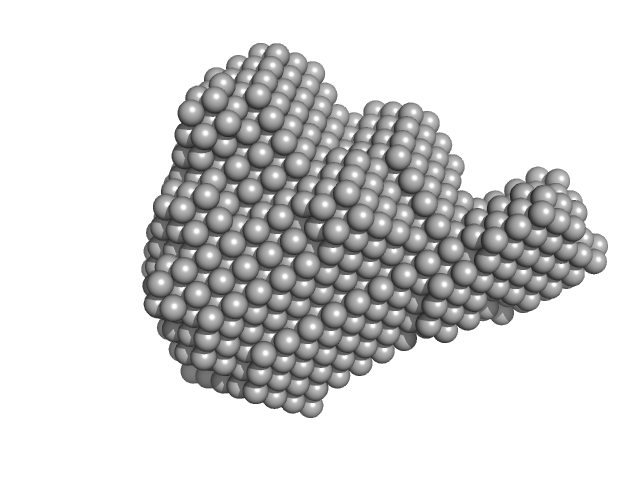
| Sample: |
Aprataxin and PNK-like factor (acidic domain) dimer, 15 kDa Homo sapiens protein
Histone H2A dimer, 26 kDa Drosophila melanogaster protein
Histone H2B dimer, 27 kDa Drosophila melanogaster protein
Histone H3 dimer, 30 kDa Drosophila melanogaster protein
Histone H4 dimer, 23 kDa Drosophila melanogaster protein
|
| Buffer: |
25 mM NaPi, 300 mM NaCl, 3% v/v glycerol, 1 mM DTT,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 10
|
Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Sci Adv 8(30):eabo0517 (2022)
Corbeski I, Guo X, Eckhardt BV, Fasci D, Wiegant W, Graewert MA, Vreeken K, Wienk H, Svergun DI, Heck AJR, van Attikum H, Boelens R, Sixma TK, Mattiroli F, van Ingen H
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
260 |
nm3 |
|
|